Abstract
Background
Rheumatoid arthritis (RA) is a chronic autoimmune disease, and understanding its genetic and molecular basis is crucial for early diagnosis, treatment, and prevention.
Objective
This study aims to explore the association between IL-4 polymorphisms (rs2227284, rs2243267, rs2243270, and rs2243283) and RA risk.
Methods
The four IL-4 polymorphisms were genotyped in 493 RA patients and 493 healthy controls using Agena MassARRAY. Logistic regression analysis calculated odds ratio (OR) and 95% confidence interval (CI) to estimate the relationship between IL-4 polymorphisms and RA risk.
Results
Overall analysis revealed that rs2243267 (GG vs. CC: OR = 0.26, FDR-p = .032; Recessive: OR = 0.27, FDR-p = .048) and rs2243270 (AA vs. GG: OR = 0.26, FDR-p = .024; Recessive: OR = 0.27, FDR-p = .024) were associated with a decreased risk of RA. Stratified analysis indicated that rs2243267 and rs2243270 were correlated with reduced RA risk in female, smoking, BMI <24, and drinking population; rs2227284 was associated with a decreased RA risk in BMI <24 and drinking population. Moreover, rs2243267 and rs2243270 were significantly associated with reduced ACPA positivity.
Conclusions
Our findings suggest that IL-4 polymorphisms (rs2227284, rs2243267, and rs2243270) act as protective factors for RA in the Chinese Han population.
1. Introduction
Rheumatoid arthritis (RA) is one of the most common chronic autoimmune diseases, characterized by synovial inflammation and joint destruction leading to tissue damage, functional impairment, severe disability, and premature mortality [Citation1,Citation2]. Account for 0.5%–1% of the peoples affected by RA worldwide, and the age-standardized prevalence and disability-adjusted life years rates of RA increased with age and were higher in females [Citation3]. RA is more common in women, who are two to three times more prone to develop the disease than men [Citation4]. Studies suggest that RA is a complex disease influenced by multiple genetic factors, environmental factors (gender, infection, and immune system), as well as gene-environment interaction [Citation5–8]. The interplay between these factors and the specific mechanisms underlying disease onset and progression are still not fully elucidated. Therefore, understanding of the genetic and molecular basis of RA will help provide better scientific basis for early diagnosis, treatment, and prevention of the disease.
Recently, studies have found that many cytokine genes variants are correlated with RA risk as cytokines play a vital role in the pathogenesis of RA [Citation9–11]. Interleukin 4 is encoded by the IL-4 gene located in chromosome 5 (q31–33), which is produced primarily by activated Th2 type CD4+ T cells, monocytes, mast cells and basophilic granulocyte. IL-4 is a cytokine with various biological functions that induces immunoglobulin E (IgE) production in B lymphocytes and serves as an important regulator of IgG isotype switching [Citation12,Citation13] and regulates the differentiations of precursor T helper cells into these of the Th2 subset that mediate humoral immunity and modulate antibody production [Citation14]. IL-4 has an immunomodulatory effect on RA, which is a small cytokine with potential for RA therapy [Citation15–17]. Numerous studies have been reported that the SNPs in IL-4 were correlated with RA risk [Citation9,Citation18,Citation19].
Despite the relationships between several SNPs in IL-4 and risk of RA has been examined, the relationships between other IL-4 polymorphisms with RA risk in the Chinese Han population remains unknown and could not be deduced. Therefore, we enrolled 493 patients with RA and 493 healthy controls to explore the relationships between four SNPs (rs2227284, rs2243267, rs2243270, and rs2243283) in IL-4 and susceptibility to RA in the Chinese Han population. Our study has the potential to significantly advance our understanding of RA pathogenesis and treatment.
2. Materials and methods
2.1. Sample size calculation and collection
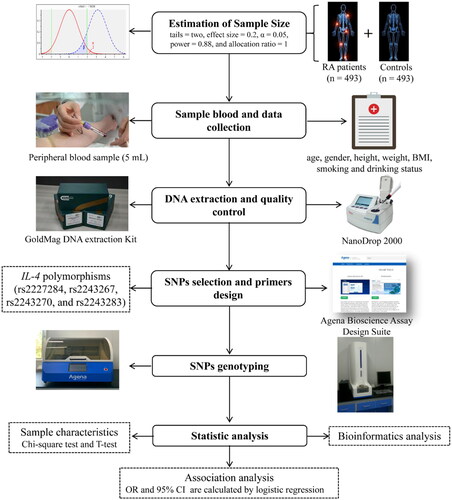
A flow diagram of this study is summarized in . The G*Power software version 3.1.9.7 was used to estimate the sample size of the case and control groups by independent samples T-test. The parameters were set as follows: tail = two, effect size = 0.23, α = 0.05, power = 0.95, allocation ratio = 1. A total of 493 patients with RA and 493 healthy controls were recruited from the Affiliated Hospital of Xizang Minzu University.
All patients were diagnosed with RA strictly according to the diagnostic criteria outlined in the 2018 Chinese Guidelines for the Diagnosis and Treatment of RA [Citation20]. No patients were included solely based on the presence of RA ICD codes; rather, a comprehensive evaluation was conducted to ensure accurate diagnosis. Specifically, patients were identified based on a combination of clinical manifestations, laboratory findings, and radiological evidence. Demographic data, including age, gender, height, weight, body mass index (BMI, kg/m2), smoking, and drinking habits, were retrieved from hospital records. For smoking status, individuals were categorized as nonsmokers if they had never smoked or smoked occasionally, while smokers were defined as those with a habitual or frequent smoking pattern. Similarly, for drinking habits, nondrinkers were classified as those who had never consumed alcohol or drank occasionally, whereas drinkers were those with a habitual or frequent drinking pattern. Additionally, laboratory testing results, such as C-reactive protein (CRP), rheumatoid factor (RF), erythrocyte sedimentation rate (ESR), anti-cyclic citrullinated peptide antibody (ACPA), and anti-Keratin antibody (AKA), were also collected for analysis.
Healthy controls were randomly recruited from the physical examination center of the Affiliated Hospital of Xizang Minzu University during the same period as the case group. To maintain homogeneity within the control group and minimize potential confounders, individuals with a history of autoimmune diseases or chronic inflammatory disease were excluded from the study. The list of excluded diseases includes, but is not limited to, systemic lupus erythematosus, chronic active hepatitis, chronic bronchitis, chronic nephritis, and other autoimmune arthritis apart from RA. Demographic information and RA risk factor data, including age, gender, height, weight, BMI, smoking, and drinking status, were collected using a pre-established questionnaire. Both patients with RA and controls are genetically unrelated Chinese Han population.
2.2. DNA extraction
Peripheral blood (5 mL) was collected from each patient with RA and control in vacutainers containing Ethylene diamine tetraacetic acid (EDTA) anti-coagulant. Genomic DNA was extracted from whole blood samples using the GoldMag-Mini Whole Blood Genomic DNA Purification Kit (GoldMag. Co. Ltd., Xi’an, China) according to the manufacturer’s specifications. The purity and concentration of the extracted DNA were tested using a Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). The concentration of qualified DNA samples should not be less than 20 ng/μl, and the DNA purity (OD260/280) should be within the range of 1.7–2.0. The DNA samples were stored at −20 °C and prepared for SNPs genotyping.
2.3. SNPs selection and genotyping
The interest gene (IL-4) was selected based on published knowledge about the importance in the immune pathway and RA development [Citation15,Citation16]. Four SNPs (rs2227284, rs2243267, rs2243270, and rs2243283) in the IL-4 gene that were not reported to be associated with RA susceptibility were selected based on previously published articles [Citation21–23], and the minor allele frequency (MAF) of these SNPs is more than 0.05 in the global population from the 1000 Genomes Project database (http://asia.ensembl.org/index.html). The potential role of SNPs was predicted using the RegulomeDB database (https://regulomedb.org/regulome-search/). The primer sequences () of polymerase chain reaction (PCR) and single base extension reaction were designed using the online software Agena Bioscience Assay Design Suite Version 2.0 (https://support.agenabio.com/s/login/?ec=302&startURL=/s/) and synthesized by Bioengineering (Shanghai Co., Ltd). The four IL-4 polymorphisms genotyping were performed using the Agena MassARRAY platform with iPLEX gold chemistry (Agena Bioscience, San Diego, CA, USA), according to the manufacturer’s instructions. The data management and analysis of genotyping results used the Agena Bioscience TYPER software (version 4.0).
Table 1. Primer sequences of PCR and unique base extension.
2.4. Statistical analysis
The distributions of continuous and categorical variables between patients with RA and healthy controls were compared using the independent sample T-test and Pearson Chi-square test, respectively. Genotype frequencies of the four SNPs (rs2227284, rs2243267, rs2243270, and rs2243283) in IL-4 in control group were assessed for Hardy-Weinberg equilibrium (HWE) using Chi-square test. The association between four SNPs in IL-4 and susceptibility to RA was evaluated based on the odds ratio (OR) and 95% confidence interval (CI) calculated by logistic regression analysis under multiple genetic models (allele, codominant, dominant, recessive, and additive) adjusted for confounding variables, including age, gender, BMI, smoking status, and drinking status. Stratified analyses were also performed using logistic regression model that were adjusted for confounding variables to minimize the impact of these factors (age, gender, BMI, smoking and drinking status) on the statistical results and thereby provide a more accurate assessment of the relationships between the SNPs in IL-4 and RA susceptibility. Forest plots of stratified results were drawn using Sangerbox software (http://sangerbox.com/home.html). To reduce false positives in the results, the p-value was corrected for error detection rate (FDR) (q = p × (n/k); N is the total number of p-values; K is the order in which p-values are sorted from minimum to maximum). All statistical analyses were performed using Microsoft Excel (Microsoft Corp., Redmond, WA, USA), Statistical Package for the Social Sciences (SPSS) version 20 (SPSS, Chicago, IL), and PLINK software (version 1.07). All statistical analyses were two sided, and the p values less than.05 were considered statistically significant.
3. Results
3.1. Characteristics of subjects
Characteristics of study subjects are presented in , including 493 patients with RA and 493 healthy controls. The patients with RA had an average age, height, weight and BMI of 54.35 ± 11.95 years, 162.03 ± 5.91 cm, 59.14 ± 8.78 kg, and 22.46 ± 2.53 kg/m2, respectively, while the control group had a mean age, height, weight and BMI of 54.02 ± 8.84 years, 162.11 ± 7.50 cm, 59.55 ± 11.34 kg, and 22.48 ± 2.74 kg/m2, respectively. T-test results indicated no significant differences in age (p = .615), height (p = .850), weight (p = .525) and BMI (p = .891) between the case and control groups. Among the male subjects, 134 (27.2%) had RA, while 124 (25.2%) were in the healthy control group. For females, 359 (72.8%) had RA and 369 (74.8%) were in the healthy control group. In addition, age and BMI were categorized based on the median age (55 years old) and Chinese BMI classification criteria (24 kg/m2). The chi-square test results revealed a well-matched distribution of gender, age and BMI, smoking, and drinking between two controls (p = .469, p = .899, p = .627, p = .792, and p = .943, respectively).
Table 2. Demographic characteristics of controls and RA patients.
3.2. Basic information and functional prediction of SNPs in IL-4
displays the basic information and allele frequency distribution of four SNPs (rs2227284, rs2243267, rs2243270, and rs2243283) in the IL-4 gene among patients with RA and healthy controls. Genotype frequency distribution of these SNPs in the IL-4 gene was found to be in accordance with HWE in the control group (p > .05) according to the chi-square test results. This indicates that the genotyping results of the SNPs are reliable, and that the selected research subjects were randomly sampled. Furthermore, based on the prediction results from the RegulomeDB database, it was found that these SNPs (rs2227284, rs2243267, and rs2243270) may influence the binding of transcription factors and subsequently impact the expression level of target genes.
Table 3. Basic information of four SNPs in the IL-4 gene.
3.3. Overall analysis of the association between SNPs in IL-4 and RA risk
We performed a genetic model analysis to investigate the associations between four SNPs (rs2227284, rs2243267, rs2243270, and rs2243283) in IL-4 with RA susceptibility, as presented in . This study revealed that carriers of the genotype GG of rs2243267 had a reduced risk of RA compared to those with the CC genotype, after adjusting for sex, age, BMI, smoking and drinking (OR = 0.26, 95% CI: 0.10–0.64, FDR-p = .032). Furthermore, rs2243267 was found to be associated with RA susceptibility under the recessive model (GG vs. CC-GC: OR = 0.27, 95% CI: 0.11–0.66, FDR-p = .048). Additionally, we observed that rs2243270 was also significantly associated with a decreased risk of RA under the codominant (AA vs. GG: OR = 0.26, 95% CI: 0.10–0.64, FDR-p = .024) and recessive models (AA vs. GG-AG: OR = 0.27, 95% CI: 0.11–0.66, FDR-p = .024).
Table 4. Association between IL-4 polymorphisms and RA risk in overall analysis.
3.4. Stratification analysis of the association between SNPs in IL-4 and RA risk
In order to reduce the potential influence of confounding factors (age, gender, BMI, smoking, and drinking status) on the results, we conducted stratification analysis. In female, rs2243267 was found to be significantly associated with reduced risk of RA (G vs. C: OR = 0.72, 95% CI: 0.56–0.94, FDR-p = .032; GG vs. CC: OR = 0.15, 95% CI: 0.04–0.53, FDR-p = .022; GG vs. CC-GC: OR = 0.16, 95% CI: 0.05–0.56, FDR-p = .022; additive: OR = 0.70, 95% CI: 0.54–0.92, FDR-p = .031, ). Meanwhile, rs2243270 was also associated with a decreased risk of RA (A vs. G: OR = 0.72, 95% CI: 0.56–0.94, FDR-p = .029; AA vs.GG: OR = 0.15, 95% CI: 0.04–0.53, FDR-p = .017; AA vs. GG-AG: OR = 0.16, 95% CI: 0.05–0.56, FDR-p = .011; additive: OR = 0.70, 95% CI: 0.54–0.92, FDR-p = .028). However, no association between the two SNPs (rs2243267 and rs2243270) and RA susceptibility in male. The other two SNPs (rs2227284 and rs2243283) in the male and female subgroups were not found to be associated with RA risk (Supplemental Table 1).
Figure 2. Forest map of the association between SNPs and RA risk in gender and smoking status stratification. RA: rheumatoid arthritis; SNP: single nucleotide polymorphism; OR: odds ratio; CI: confidence interval; FDR: false discovery rate. p < .05 was considered to be significant.
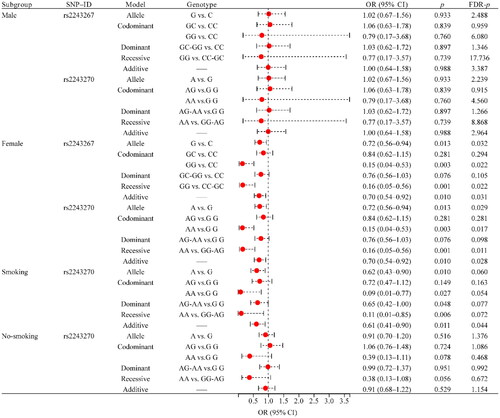
In addition, we found that rs2243270 (OR = 0.61, 95% CI: 0.41–0.90, FDR-p = .044) was associated with reduced RA risk in the smoking subgroup under the additive model, while no correlation was found with RA risk in the nonsmoking population (). The other three SNPs (rs2227284, rs2243267, and rs2243283) in the smoking and nonsmoking subgroups were not found to be associated with RA risk (Supplemental Table 1). We also conducted a stratified analysis of age based on the median and found that no correlation between these four SNPs and RA risk was found in both subgroups age ≥55 and <55 (Supplemental Table 1).
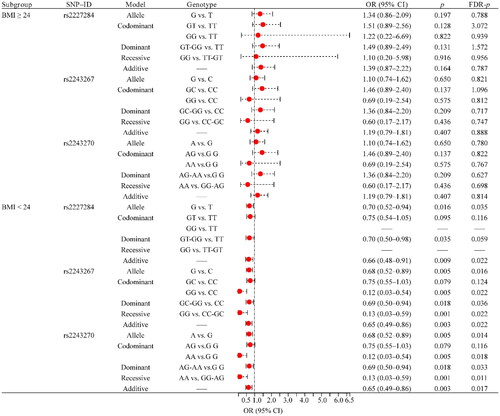
The BMI stratification results indicated that rs2227284 (G vs. T: OR = 0.70, 95% CI: 0.52–0.94, FDR-p = .035; additive: OR = 0.66, 95% CI: 0.48–0.91, FDR-p = .022), rs2243267 (G vs. C: OR = 0.68, 95% CI: 0.52–0.89, FDR-p = .016; GG vs. CC: OR = 0.12, 95% CI: 0.03–0.54, FDR-p = .022; GC-GG vs. CC: OR = 0.69, 95% CI: 0.50–0.94, FDR-p = .036; GG vs. CC-GC: OR = 0.13, 95% CI: 0.03–0.59, FDR-p = .022; additive: OR = 0.65, 95% CI: 0.49–0.86, FDR-p = .022), rs2243270 (A vs. G: OR = 0.68, 95% CI: 0.52–0.89, FDR-p = .014; AA vs. G G: OR = 0.12, 95% CI: 0.03–0.54, FDR-p = .018; AG-AA vs.GG: OR = 0.69, 95% CI: 0.50–0.94, FDR-p = .033; AA vs. GG-AG: OR = 0.13, 95% CI: 0.03–0.59, FDR-p = .011; additive: OR = 0.65, 95% CI: 0.49–0.86, FDR-p = .017) was associated with RA susceptibility in BMI <24 (). However, no association between these SNPs and RA susceptibility was found in the BMI ≥ 24 subgroup ( and Supplemental Table 1).
Figure 3. Forest map of the association between SNPs and RA risk in BMI stratification. RA: rheumatoid arthritis; BMI: body mass index; SNP: single nucleotide polymorphism; OR: odds ratio; CI: confidence interval; FDR: false discovery rate. p < .05 was considered to be significant.
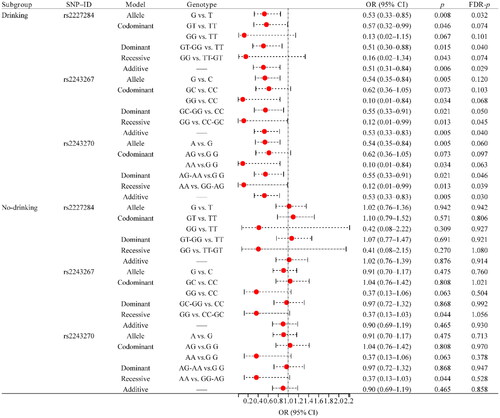
The drinking status stratification results showed that rs2227284 (G vs. T: OR = 0.53, 95% CI: 0.33–0.85, FDR-p = .032; GT-GG vs. TT: OR = 0.51, 95% CI: 0.30–0.88, FDR-p = .040; additive: OR = 0.51, 95% CI: 0.31–0.84, FDR-p = .029), rs2243267 (GG vs. CC-GC: OR = 0.12, 95% CI: 0.01–0.99, FDR-p = .045; additive: OR = 0.53, 95% CI: 0.33–0.83, FDR-p = .040), and rs2243270 (AG-AA vs. G G: OR = 0.55, 95% CI: 0.33–0.91, FDR-p = .046; AA vs. GG-AG: OR = 0.12, 95% CI: 0.01–0.99, FDR-p = .039; additive: OR = 0.53, 95% CI: 0.33–0.83, FDR-p = .030) in smoking population (). However, no association between these SNPs and RA susceptibility was found in the nonsmoking subgroup ( and Supplemental Table 1).
3.5. Association between SNPs in IL-4 and clinical indicators
To further explore the association between IL-4 polymorphisms and RA risk, we analyzed the correlation between SNPs (rs2227284, rs2243267, rs2243270, and rs2243283) in IL-4 and clinical indicators (CRP, RF, ESR, ACPA, and AKA) of RA patients. The results showed that rs2243267 (OR = 0.07, 95% CI: 0.01–0.39, p = .002) and rs2243270 (OR = 0.07, 95% CI: 0.01–0.39, p = .002) were significantly correlated with a decrease in ACPA positivity ().
Table 5. Association between IL-4 polymorphisms and clinical indicators of RA patients.
4. Discussion
We conducted a case-control study comprising 493 RA patients and 493 healthy controls to investigate the influence of other IL-4 polymorphisms (rs2227284, rs2243267, rs2243270, and rs2243283) on the susceptibility to RA in the Chinese Han population. The overall analysis revealed that two SNPs (rs2243267 and rs2243270) in IL-4 were correlated with a decreased risk of RA. Stratification analysis results indicated that these two SNPs were significantly associated with reduced risk of RA in female, smoking, BMI <24, and drinking subgroups. Meanwhile, rs2227284 was associated with RA susceptibility in BMI <24 and drinking subgroups; and rs2243283 was found to be associated with increased risk of RA in smoking subgroups. Moreover, rs2243267 and rs2243270 were significantly associated with reduced ACPA positivity.
IL-4 is a major effector cytokine produced by activated CD4 + T cells. It promotes the polarization of Th2 cells in vitro, which was originally identified as B-cell stimulating factor and regulates isotype class switching of B cells toward IgG1 and IgE [Citation24,Citation25]. It has been reported that IL-4 exerts great anti-inflammatory role in RA [Citation26]. Previous studies have reported that IL-4 polymorphisms were significantly associated with susceptibility to many diseases. For example, rs2227284 was significantly associated with susceptibility to many diseases, such as lung cancer [Citation23], colorectal cancer [Citation27], allergic rhinitis [Citation28,Citation29]. The SNP (rs2227284) was found obviously associated with renal cell carcinoma (RCC) risk in nonsmoking and drinking subgroup [Citation21]. Our study is the first to find a significant association between rs2227284 and RA risk in overall, BMI <24 and smoking subgroups. The two IL-4 polymorphisms (rs2243267 and rs2243270) were found obviously associated with RCC in age < 55 and drinking subgroup [Citation21]. In this study, rs2243267 and rs2243270 were significantly associated with reduced risk of RA in overall, female, BMI <24, drinking, and smoking subgroups. Furthermore, rs2243283 was reported to be associated with risk of steroid-induced osteonecrosis of the femoral head (ONFH) [Citation22], esophageal squamous cell carcinoma [Citation30], and idiopathic-nephrotic syndrome [Citation31]. This study did not find a significant association between rs2243283 and risk of RA.
Previous studies have confirmed gender differences in the epidemiological and clinical manifestations of RA, and IL-4 may be a potential gene responsible for the pathogenesis of RA and leads to differences in RA between women and men [Citation32]. Smoking has been implicated as one of the most important extrinsic risk factors for RA development and severity [Citation33]. A prospective Chinese cohort study found that increasing alcohol consumption was associated with an increased risk of RA in women [Citation34]. A National Health and Nutrition Examination Survey (NHANES) and meta-analysis in USA showed that nearly 33% of RA incidence was attributed to smoking, high BMI and low alcohol consumption [Citation35]. These findings of this study stratified analysis may suggest that RA genetic susceptibility varies by sex, age, BMI, smoking, and drinking status, and highlight the importance of considering heterogeneity in future genetic association studies of RA risk. Therefore, a deeper study of the interaction between stratification factors and genetic susceptibility to RA will help predict and prevent the development of this disease more accurately. Future research needs to further explore the impact of these factors on the genetic susceptibility of RA, in order to improve our understanding of its pathogenesis and provide a basis for developing more personalized treatment strategies. In addition, these results also remind people to reduce the risk of RA through lifestyle interventions, such as weight control, smoking cessation, and alcohol restriction.
CRP is routinely assessed as a marker of systemic inflammation in RA [Citation36]. RF is a well-established marker for the diagnosis and classification of RA [Citation37]. AKA has high diagnostic specificity in RA and may be useful for RA diagnostic application in clinic [Citation38]. Previous study shown that individuals with high ACPA titers have an increased risk of developing RA [Citation39]. In this study, rs2243267 and rs2243270 were found to be associated with a decreased ACPA positive. Therefore, we speculate that these two SNPs may reduce the risk of RA by reducing the ACPA positivity rate. However, it is necessary to further explore the role of these SNPs in the occurrence and development of RA.
In summary, the present study exhibits several key advantages. First, its robust case-control design significantly enhances the reliability and validity of our findings. Second, by focusing exclusively on the genetically homogenous Chinese Han population, the study effectively mitigates the confounding effects of ethnic and genetic diversity, allowing for a more precise assessment of the association between IL-4 SNPs and RA susceptibility. Furthermore, the study’s stratification analysis, which takes into account various potential confounding factors such as gender, smoking status, BMI, and drinking habits, enables us to identify subgroup-specific associations, thereby providing a more nuanced understanding of the complex relationship between IL-4 SNPs and RA risk. Finally, our study goes beyond merely examining the association between IL-4 SNPs and RA risk, exploring their correlation with ACPA positivity, a crucial biomarker in RA. This added dimension to our analysis offers valuable insights into the potential mechanistic roles of these SNPs in RA pathogenesis, further strengthening the impact and significance of our research.
Despite the potential significance of our study in exploring the association between IL-4 gene polymorphisms and RA susceptibility in the Chinese Han population, there are several limitations that must be considered. First, our study focused exclusively on the Chinese Han population and may not be generalizable to other ethnic groups or populations with different genetic and environmental backgrounds. Second, we only randomly selected four SNPs in the IL-4 gene, and did not explore the association between more IL-4 polymorphisms and RA risk. Finally, we only speculated about the reason why IL-4 polymorphisms regulates RA risk, but did not conduct functional verification. Therefore, it is necessary to further validate the results of this study and explore the mechanism of IL-4 polymorphisms in RA. Through continued research on the molecular mechanisms underlying RA susceptibility, we can better understand this complex disease and ultimately develop more effective treatments and prevention strategies for those affected.
5. Conclusion
In conclusion, our results suggested that four IL-4 polymorphisms (rs2243267, rs2243270, and rs2243283) ware associated with decrease risk of RA in Chinese Han population. However, this study is a pilot study and further studies with large samples are warranted to confirm the results and evaluate the significance of our findings in the clinic.
Author’s contributions
Li Wang responsible for the study design. Xiaoli Liu and Huqiang Mai contributed to manuscript preparation and writing. Liang Wang performed the experiments and collected the samples. Hengxun Zhang contributed to data analysis and interpretation. Xuemei Li and Xuguang Li were responsible for the manuscript revision. All authors were responsible for drafting the manuscript, read and approved the final version.
Ethical approval
This study was approved by the Ethics Committee of the Affiliated Hospital of Xizang Minzu University (No. 20190-5) and was conducted according to the principles of the Declaration of Helsinki. All the participants signed written informed consents for blood samples collection and subsequent analysis at recruitment.
Consent for publication
Not applicable
Supplemental Material
Download PDF (217.8 KB)Acknowledgements
The authors thank all participants for their support and participation.
Disclosure statement
The authors declare that they have no competing interests in this work.
Data availability statement
The datasets generated and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Additional information
Funding
References
- Aletaha D, Smolen JS. Diagnosis and management of rheumatoid arthritis: a review. JAMA. 2018;320(13):1–11.
- Sparks JA. Rheumatoid arthritis. Ann Intern Med. 2019;170(1):ITC1–ITC16.
- Safiri S, Kolahi AA, Hoy D, et al. Global, regional and national burden of rheumatoid arthritis 1990-2017: a systematic analysis of the Global Burden of Disease study 2017. Ann Rheum Dis. 2019;78(11):1463–1471.
- Favalli EG, Biggioggero M, Crotti C, et al. Sex and management of rheumatoid arthritis. Clin Rev Allergy Immunol. 2019;56(3):333–345.
- Weyand CM, Goronzy JJ. The immunology of rheumatoid arthritis. Nat Immunol. 2021;22(1):10–18.
- Atzeni F, Masala IF, di Franco M, et al. Infections in rheumatoid arthritis. Curr Opin Rheumatol. 2017;29(4):323–330.
- Klareskog L, Rönnelid J, Saevarsdottir S, et al. The importance of differences: On environment and its interactions with genes and immunity in the causation of rheumatoid arthritis. J Intern Med. 2020;287(5):514–533.
- Padyukov L. Genetics of rheumatoid arthritis. Semin Immunopathol. 2022;44(1):47–62.
- Yucel B, Sumer C, Gok I, et al. Associations between cytokine gene polymorphisms and rheumatoid arthritis in Turkish population. North Clin Istanb. 2020;7(6):563–571.
- Agonia I, Couras J, Cunha A, et al. IL-17, IL-21 and IL-22 polymorphisms in rheumatoid arthritis: a systematic review and meta-analysis. Cytokine. 2020;125:154813.
- Xie Q, Xu W-D, Pan M, et al. Association of IL-35 expression and gene polymorphisms in rheumatoid arthritis. Int Immunopharmacol. 2021;90:107231.
- Brown MA, Hural J. Functions of IL-4 and control of its expression. Crit Rev Immunol. 2017;37(2-6):181–212.
- Junttila IS. Tuning the cytokine responses: an update on interleukin (IL)-4 and IL-13 receptor complexes. Front Immunol. 2018;9:888.
- Kubo M. The role of IL-4 derived from follicular helper T (TFH) cells and type 2 helper T (TH2) cells. Int Immunol. 2021;33(12):717–722.
- Dong C, Fu T, Ji J, et al. The role of interleukin-4 in rheumatic diseases. Clin Exp Pharmacol Physiol. 2018;45(8):747–754.
- Chen Z, Bozec A, Ramming A, et al. Anti-inflammatory and immune-regulatory cytokines in rheumatoid arthritis. Nat Rev Rheumatol. 2019;15(1):9–17.
- Spieler V, Ludwig MG, Dawson J, et al. Targeting interleukin-4 to the arthritic joint. J Control Release. 2020;326:172–180.
- Sun YH, Wei ST, Zong SH. Correlation between IL-4 gene polymorphismas well as its mRNA expressionand rheumatoid arthritis. Eur Rev Med Pharmacol Sci. 2017;21(17):3879–3885.
- Elshazli RM, Elsaid AM, Shawky DM, et al. Genetic polymorphisms of ACE I/D, IL-1beta G > A and IL-4 VNTR among Egyptian subjects with rheumatoid arthritis. Arch Physiol Biochem. 2022; 128(3):576–585.
- Tian X, Wang Q, Li M, et al. 2018 Chinese guidelines for the diagnosis and treatment of rheumatoid arthritis. Rheumatol Immunol Res. 2021;2(1):1–14.
- Rong H, He X, Wang L, et al. Associations between polymorphisms in the IL-4 gene and renal cell carcinoma in Chinese Han population. Oncotarget. 2017;8(47):82078–82084.
- Jin T, Zhang Y, Sun Y, et al. IL-4 gene polymorphisms and their relation to steroid-induced osteonecrosis of the femoral head in Chinese population. Mol Genet Genomic Med. 2019;7(3):e563.
- Tan N, Song J, Yan M, et al. Association between IL-4 tagging single nucleotide polymorphisms and the risk of lung cancer in China. Mol Genet Genomic Med. 2019;7(4):e00585.
- Zhu J. T helper 2 (Th2) cell differentiation, type 2 innate lymphoid cell (ILC2) development and regulation of interleukin-4 (IL-4) and IL-13 production. Cytokine. 2015;75(1):14–24.
- Yoshimoto T. The hunt for the source of primary interleukin-4: how we discovered that natural killer T cells and basophils determine T helper type 2 cell differentiation in vivo. Front Immunol. 2018;9:716.
- Iwaszko M, Biały S, Bogunia-Kubik K. Significance of interleukin (IL)-4 and IL-13 in inflammatory arthritis. Cells. 2021;10(11):3000.
- Wen X, Xin X, Li J, et al. The correlation between IL-4 polymorphisms and colorectal cancer risk in a population in Northwest China. Eur J Cancer Prev. 2020;29(2):95–99.
- Li Y, Chen J, Rui X, et al. The association between sixteen genome-wide association studies-related allergic diseases loci and childhood allergic rhinitis in a Chinese Han population. Cytokine. 2018;111:162–170.
- Moh’d Al-Rawashdeh B, Sada Alhanjori A, Ali E, et al. Association of IL-4 polymorphisms with allergic rhinitis in Jordanian population. Medicina (Kaunas). 2020;56(4):56.
- Wang J, Chen T, Tang W, et al. Associations of interleukin-4 and interleukin-4 receptor loci with esophageal squamous cell carcinoma susceptibility. Int Immunopharmacol. 2021;97:107659.
- Li W, Li L, He L, et al. Cytokine gene polymorphisms in Chinese children with idiopathic nephrotic syndrome. Iran J Immunol. 2022;19(1):9.
- Yu C, Liu C, Jiang J, et al. Gender differences in rheumatoid arthritis: interleukin-4 plays an important role. J Immunol Res. 2020;2020:4121524.
- Chang K, Yang SM, Kim SH, et al. Smoking and rheumatoid arthritis. Int J Mol Sci. 2014;15(12):22279–22295.
- VanEvery H, Yang W, Olsen N, et al. Alcohol consumption and risk of rheumatoid arthritis among Chinese adults: a prospective study. Nutrients. 2021;13(7):13.
- Ye D, Mao Y, Xu Y, et al. Lifestyle factors associated with incidence of rheumatoid arthritis in US adults: analysis of National Health and Nutrition Examination Survey database and meta-analysis. BMJ Open. 2021;11(1):e038137.
- Pope JE, Choy EH. C-reactive protein and implications in rheumatoid arthritis and associated comorbidities. Semin Arthritis Rheum. 2021;51(1):219–229.
- Van Hoovels L, Vander Cruyssen B, Sieghart D, et al. IgA rheumatoid factor in rheumatoid arthritis. Clin Chem Lab Med. 2022;60(10):1617–1626.
- Wang X-P, Cheng Q-Y, Gu M-M, et al. Diagnostic accuracy of anti-keratin antibody for rheumatoid arthritis: a meta-analysis. Clin Rheumatol. 2019;38(7):1841–1849.
- Wu CY, Yang HY, Lai JH. Anti-citrullinated protein antibodies in patients with rheumatoid arthritis: biological effects and mechanisms of immunopathogenesis. Int J Mol Sci. 2020;21(11):4015.