Abstract
Sessile cultures of the skin bacteria Staphylococcus saprophyticus and Corynebacterium xerosis were grown using novel fine-celled foam substrata to test the outcome of challenge by methicillin-resistant Staphylococcus aureus or Pseudomonas aeruginosa under three growth medium regimens (simulated sweat, simulated serum or simulated sweat substituted with simulated serum during the microbial challenge). S. saprophyticus and C. xerosis significantly limited MRSA and P. aeruginosa immigration respectively, under the simulated sweat and serum medium regimes. Under the substitution medium regime however, MRSA and P. aeruginosa integrated into pre-established biofilms to a significantly greater extent, attaining cell densities similar to the axenic controls. The outcome of challenge was influenced by the medium composition and test organism but could not be predicted based on planktonic competition assays or growth dynamics. Interactions between skin and wound isolates could be modelled using the fine-celled foam-based system. This model could be used to further investigate interactions and also in preclinical studies of antimicrobial wound care regimens.
Introduction
In vitro biofilm models have applications ranging from mechanistic studies of biofilm-specific processes to preclinical investigations of antimicrobial products (reviewed by McBain Citation2009). Various biofilm models have been developed including the constant depth film fermenter (CDFF) in which continuous biofilm cultures of pre-set depth are grown within coupons housed in a rotating turntable (Kinniment et al. Citation1996; Pratten et al. Citation1998; McBain et al. Citation2003a; Ledder et al. Citation2009). The CDFF has most frequently been used to maintain oral consortia but also to grow mixed populations of wound bacteria (Malic et al. Citation2009; Hill et al. Citation2010), for other applications including the growth of aquatic ecosystems (McBain et al. Citation2004) and to model interactions between Burkholderia cepacia and Pseudomonas aeruginosa of relevance to cystic fibrosis (Al-Bakri et al. Citation2004). Whilst the CDFF has proven utility, the rotary action of the turntable and static scraper blades of the CDFF create shear forces that are unlikely to be commonly encountered in wounds. Various other systems, all with inherent strengths and weaknesses, have been adopted to model wound biofilms, including the CDC reactor (Humphreys et al. Citation2011), porcine dermal explants (Barnea et al. Citation2010; Yang et al. Citation2013), microfluidics (Wright et al. Citation2015), collagen-based systems (Charles et al. Citation2009) and human skin equivalents generated from primary human keratinocytes (Haisma et al. Citation2013).
Typically, in vitro wound biofilm models utilise immobilised abiotic surfaces in either continuously perfused or batch culture systems. Perfused models including the single (Taylor & Greenman Citation2010) and multiple Sorbarod devices (Ledder et al. Citation2009), modified drip slide reactors (Agostinho et al. Citation2011; Woods et al. Citation2012), and a specialised perfused biofilm model described by Thorn and Greenman (Citation2009), incorporate porous substrata and have been used to grow biofilms compositionally similar to those occurring in wounds. Static or batch culture systems such as the Lubbock chronic wound model (Dowd et al. Citation2008; Sun et al. Citation2008), the modified Lubbock model described by Kucera et al. (Citation2014) and a cellulose disc model described by Hammond et al. (Citation2011) have been used to investigate multispecies wound biofilms and antimicrobial dressings to good effect.
In the current investigation, mono- and binary culture biofilms were grown using water-absorbing thermoset plastic foam (Smithers-Oasis, Washington, UK) marketed for the postharvest maintenance of plants, as substrata. Whilst in vitro models have been used to investigate aspects of biofilms formed by bacteria that colonise wounds, particularly in terms of the effect of antimicrobial compounds and dressings (Steffansen & Herping Citation2008; Sun et al. Citation2008; Thorn & Greenman Citation2009; Lipp et al. Citation2010), interactions between populations of microorganisms associated with intact skin and with dermal wounds have received relatively little research attention. With the broad aim of ascertaining whether growth interactions between bacteria representing skin commensals and wound pathogens could be investigated using a simple in vitro system, a fine-celled foam model was used to simulate the challenge of established populations of coagulase-negative staphylococci and C. xerosis with the adventitious pathogens methicillin-resistant Staphylococcus aureus (MRSA) and P. aeruginosa, under growth substrate conditions broadly reflective of intact or wounded skin.
Materials and methods
Bacteria
P. aeruginosa and C. xerosis were isolated from infected wounds; methicillin resistant S. aureus NCTC 11939 and S. saprophyticus NCTC 7292 were obtained from Public Health England, Southampton, UK.
Chemicals and media
Unless otherwise stated chemicals used were supplied by Sigma (Poole, UK). Dehydrated bacteriological media were obtained from Oxoid (Basingstoke, UK) and reconstituted according to instructions supplied by the manufacturer.
Simulated sweat and serum formulations
To simulate and standardise the nutrients typically available to bacteria on intact and wounded skin, growth media broadly reflective of the major constituents of hominine sweat and serum at biologically relevant concentrations were developed (Russell & Wiles Citation1970; Trengove & Langton Citation1996). The simulated sweat consisted of 3-(N-morpholino)-propanesulfonic acid (20.9 g l−1/100 mM), yeast extract (1 g l−1), NaCl (2 g l−1), fish oil fatty acid methyl esters (0.65 mg l−1) and Tween 80 (0.1 g l−1) buffered to pH 6. Simulated serum consisted of MOPS (20.9 g l−1/100 mM), NaCl (6.025 g l−1), KCl (0.372 g l−1), urea (0.54 g l−1), creatinine (0.0132 g l−1), glucose (0.324 g l−1), yeast extract (1 g l−1), peptone (3 g l−1), MgSO4 (0.0168 g l−1), haemin (0.005 g l−1) and K3PO4 (0.109 g l−1).
Differential enumeration of bacteria
For mixed cultures of Staphylococcus saprophyticus and MRSA, bacteria were enumerated using Wilkins-Chalgren agar supplemented with novobiocin (5 mg l−1) or cefoxitin (4 mg l−1), respectively; for mixed cultures of S. saprophyticus and P. aeruginosa, Mannitol Salt and Pseudomonas-selective agars were used. For cultures of C. xerosis combined with MRSA, bacteria were enumerated using Wilkins-Chalgren agar and Wilkins-Chalgren agar supplemented with cefoxitin. For mixed cultures of C. xerosis and P. aeruginosa, Wilkins-Chalgren agar and Pseudomonas-selective agar facilitated selective enumeration. Viable counts of C. xerosis in combined culture were determined from the total counts derived from Wilkins-Chalgren agar, minus selective counts.
Bacterial specific growth rates and productivity
Stationary phase cultures of the skin and wound-associated bacterial isolates were then diluted 1:100 in fresh media and 200 μl of the inocula were dispensed into wells of a microtitre plate. Microtitre plates were incubated in an automated plate reader (Titertek Multiskan® MCC 340, Biotek, Swindon, UK) at 37°C for 24 h with optical density readings (600 nm) taken every 20 min. Specific growth rates and delta OD values were determined by the following equations (Inniss & Mayfield Citation1978) where Χ1 and X2 are the OD600 values on the maximum of the slope of growth curves between times t1 and t2.
Relative fitness of combined planktonic cultures
The relative fitness of planktonic cultures of S. saprophyticus or C. xerosis in pair-wise combination with either MRSA or P. aeruginosa was assessed using a competitive fitness assay previously described by Lenski et al. (Citation1991) and Rozen et al. (Citation2007). Briefly, stationary phase axenic bacterial cultures maintained in simulated sweat or serum were adjusted to ~8.0 log10 CFU ml−1 in sterile medium. Pair-wise combinations were mixed in 50:50 ratios and further adjusted to give final inoculum density of ~6.0 log10 CFU ml−1 of each bacterial isolate. Mixed inocula were serially diluted to verify the initial population densities and were then incubated for 24 h at 37°C. To determine the endpoint densities the mixed inocula were serially diluted and viable counts performed. All pair-wise combinations were tested in triplicate.
Use of a fine-celled foam multi-well plate wound model to investigate community integration and colonisation resistance
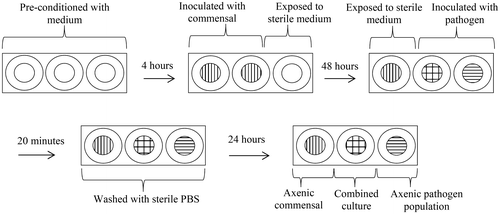
To investigate interactions between established and introduced bacteria, a fine-celled foam (FCF) multi-well model was used. A fine celled thermoset phenolic plastic foam (Smithers-Oasis) was selected as a model substratum. This foam consists of a cellular solid structure with interconnecting pores, which drives medium uptake by capillary action. It is structurally stable when saturated and has excellent wet heat stability facilitating sterilisation by autoclaving. Sterile FCF portions were cut to column size (height 1 cm × 1 cm diameter) and placed within the wells of a sterile 24 multi-well culture plate (Sigma). An overview of the inoculation procedure and incubation steps is shown in Figure . Briefly FCF substrata were pre-conditioned in simulated media for 4 h and then either inoculated with 1.0 ml stationary phase cultures of S. saprophyticus or C. xerosis (adjusted to ~7.0 log10 CFU ml−1) or left as uninoculated controls. The FCF substrata were then incubated aerobically at 37°C. After 48 h incubation, inoculated and uninoculated substrata were exposed to 1 ml of ~7.0 log10 CFU ml−1 of MRSA or P. aeruginosa, with the remaining inoculated substrata exposed to 1 ml of sterile medium for 20 min. All FCF substrata were then immersed in sterile PBS (pH 7.0, 0.01 M), and returned to their original wells containing fresh sterile medium. After further incubation for 24 h, the FCF substrata were aseptically removed and placed into plastic Universal bottles (Scientific Laboratory Supplies, Nottingham, UK) containing 9 ml of half-strength thiogylcolate broth and 5 mm sterile glass beads (n = 5) (Merck, Darmstadt, Germany). To ensure the disintegration of the substrata, effective extraction of bacterial cells, and uniform distribution of cells throughout the diluent, the Universal bottles and contents were mixed at 800 rpm on a reciprocal flask shaker (Griffin & George, Loughborough, UK) for 2 min. Extracted cells were then serially diluted in half-strength thiogylcolate broth and plated on variously selective agars, as outlined above (McBain et al. Citation2003b; Matejka et al. Citation2012). All substrata were maintained under fed-batch conditions with spent medium removed and replaced with fresh medium every 24 h. All experiments were undertaken in triplicate and repeated under three growth medium regimens: (1) simulated sweat; (2) simulated serum; or (3) simulated sweat with medium switched to simulated serum during the microbial challenge. Statistical significance was determined using independent t-test and Mann–Whitney test to determine significant difference.
Results
Bacterial specific growth rates and productivity in formulated media
Both simulated sweat and serum sustained the growth of the test bacteria although growth rates and productivity for both MRSA (0.201 and 0.838 h−1) and P. aeruginosa (0.136 and 0.833 h−1) were markedly higher than for the cutaneous organisms C. xerosis (0.098 and 0.610 h−1) and S. saprophyticus (0.106 and 0.470 h−1) in simulated sweat and serum, respectively (Table ).
Table 1. Specific growth rate and delta OD values of organisms grown in formulated simulated sweat.
Relative fitness of combined planktonic cultures
When grown together in binary planktonic culture, MRSA and P. aeruginosa were significantly fitter than C. xerosis and S. saprophyticus in both the simulated serum and sweat (Table ).
Table 2. Relative fitness of selected organisms when combined in co-culture.
Bacterial integration and colonisation resistance
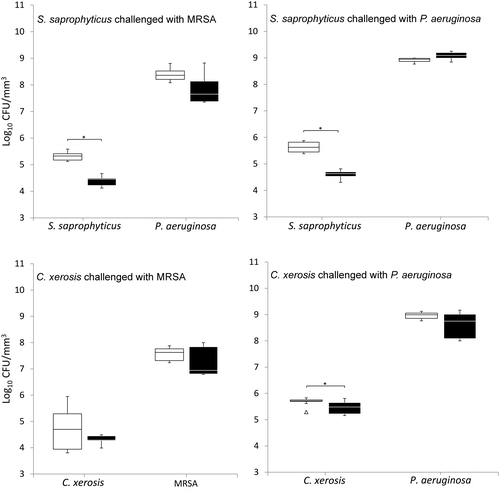
When grown in simulated sweat or serum and compared to non-colonised controls, prior colonisation of S. saprophyticus and C. xerosis resulted in significantly reduced integration of MRSA and P. aeruginosa on FCF substrata, as shown in Figures and (p < 0.05) in five of eight cases, with the exception of S. saprophyticus when challenged with P. aeruginosa under simulated serum and C. xerosis when challenged with MRSA under simulated serum and sweat. Significant displacement of the pre-colonised organisms also occurred (p < 0.05) with viable counts of C. xerosis significantly lower (p < 0.05) than for axenic controls when challenged with MRSA under simulated sweat feeding and when challenged with P. aeruginosa under both simulated sweat and serum (Figures and ). Significantly lower densities of S. saprophyticus were detected when challenged with P. aeruginosa in simulated sweat (Figure ) and when challenged by either pathogen under simulated serum (Figure ). Under the substitution nutrient regime, both MRSA and P. aeruginosa successfully integrated into established cultures of S. saprophyticus and C. xerosis, producing population densities that were not significantly different from their axenic controls (Figure ). To check for the presence of sessile bacteria and biofilms within the FCF material, selected samples were visualised using environmental scanning electron microscopy. According to this, putative attached microcolonies were present on the surfaces of inoculated FCF substrata that were absent on the control material (images not shown).
Figure 2. Viable counts of axenic and combined cultures of pre-established communities of S. saprophyticus and C. xerosis when exposed to the transient pathogenic bacteria MRSA and P. aeruginosa in simulated sweat. Data are means of three separate experiments. Asterisks indicate significant differences (p < 0.05). White boxes, axenic culture; black boxes, binary culture; minimum and maximum outliers are also indicated.
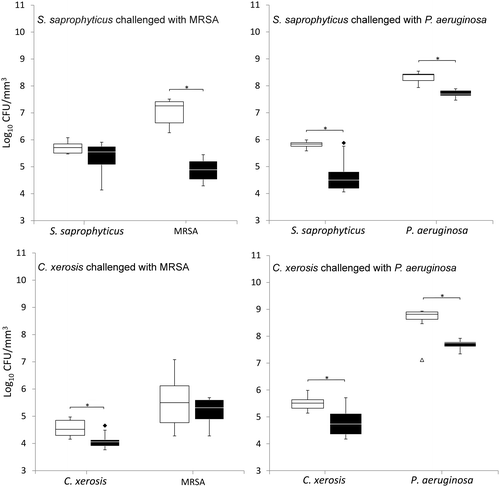
Figure 3. Viable counts of axenic and combined cultures of pre-established communities of S. saprophyticus and C. xerosis when exposed to the transient pathogenic bacteria MRSA and P. aeruginosa in simulated serum. See legend to Figure .
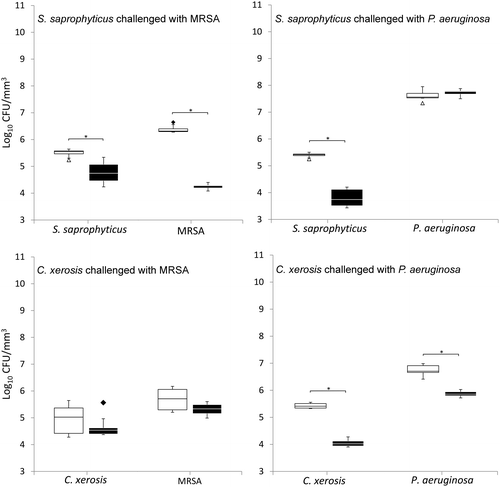
Figure 4. Viable counts of axenic and combined cultures of pre-established communities of S. saprophyticus and C. xerosis when exposed to the transient pathogenic bacteria MRSA and P. aeruginosa in simulated sweat with medium substituted with simulated serum during microbial challenge. See legend to Figure .
Discussion
The presence (Malic et al. Citation2009; Oates et al. Citation2014) and purported role biofilms play in wounds (Bjarnsholt et al. Citation2008; James et al. Citation2008) has been the focus of considerable attention in wound healing research and there is substantial commercial interest in developing dressings and combinations of antimicrobials with enhanced anti-biofilm activity (Kostenko et al. Citation2010; Kucera et al. Citation2014). Various in vitro models have been used to study wound biofilms (Sun et al. Citation2008; Thorn & Greenman Citation2009; Dalton et al. Citation2011; Kucera et al. Citation2014) and to facilitate the preclinical assessment of potential treatment outcomes (Kostenko et al. Citation2010; Agostinho et al. Citation2011; Woods et al. Citation2012). Interactions between pathogens associated with wounds and sessile populations of ‘commensal’ organisms have however received relatively little research attention. As with other microbial communities associated with the human body where microbially mediated colonisation resistance is likely to be an important protective process, it is believed that the skin microbiota, although of comparatively low density (normally ranging between 102 CFU cm−2 and 107 CFU cm−2; Fredricks Citation2001), can inhibit the attachment and proliferation (Axelsson & Mahida Citation2000) of adventitious pathogens via competition for attachment sites, nutrients and the production of inhibitory metabolites such as lactic acid and antimicrobial molecules such as bacteriocins (McAuliffe et al. Citation2001; Varella Coelho et al. Citation2007).
With the aim of using a biofilm model based on a fine-celled foam substratum to investigate the outcome of binary bacterial interactions of the type that may occur on the skin in health and during wounding, pair-wise interactions between two organisms typically isolated from the epidermis (S. saprophyticus and C. xerosis) or associated with wound infections (MRSA and P. aeruginosa) were studied under growth substrate conditions broadly reflective of intact or wounded skin using a biofilm model based on fine-celled foam. The organisms were selected based on their common isolation from either the epidermis or wounds, combined with their antibiograms and colony morphology on selective agar plates, which facilitated selective growth of individual organisms when grown in defined consortia.
The capacity of the test bacteria to grow in the formulated media was evaluated and all grew in both simulated serum and sweat although considerably higher growth rates and bacterial productivity occurred for the wound pathogens. This was reflected in the outcome of pair-wise growth interactions in relative fitness assays in which ratios of the growth rates of each organism in co-culture were determined. When planktonic cultures of MRSA or P. aeruginosa were combined with either S. saprophyticus or C. xerosis, significant advantageous growth for both pathogens occurred. As no antagonistic activity between these organisms was detected using deffered or direct antagonism assays (data not shown), this suggests that the competitiveness of the pathogenic species was most likely due to relative growth rates rather than antagonism. The utility of a fine-celled foam model for investigations of bacterial community integration, displacement or colonisation resistance and the influence of variable nutritional conditions on these factors was then assessed.
In the majority of previous reports into the use of in vitro models to simulate aspects of the bacteriology of human skin and wounds, a range of model systems have been used mainly to assess the attachment and biofilm formation by bacteria associated with wounds (Sun et al. Citation2008; Malic et al. Citation2009; Thorn & Greenman Citation2009), their responses to antibiotics (Gander et al. 2002) or to test topical antimicrobials (Dowd et al. Citation2009; Lipp et al. Citation2010). The current investigation differs from some previous reports in that it assesses the use of a biofilm model system to investigate interactions between bacteria. A novel substratum material was used in 24-well plate system, thus facilitating biological replication, and nutrient conditions were varied in order to broadly simulate conditions associated with healthy skin (simulated sweat medium) and chronic wounds (simulated serum) and through the use of varied nutrient conditions to represent wounding (medium substitution) intended to simulate changes in nutrient availability likely to occur during wounding, from the nutrient-limited environment of the skin to the more nutritious environment of a wound. During the validation phase of the study, inoculated FCF substrata were imaged using environmental scanning electron microscopy to determine whether biofilms were present within the substrata. Structures resembling attached biofilm microcolonies were observed on inoculated substrata, but not on uninoculated controls.
Under the simulated sweat and serum regimes, limited integration of the wound isolates into established populations of either S. saprophyticus or C. xerosis occurred, an outcome which could not be predicted based on planktonic pair-wise competition assays or planktonic growth dynamics, where both MRSA and P. aeruginosa consistently outcompeted S. saprophyticus and C. xerosis when grown in simulated sweat or serum. Interestingly, under the medium substitution regime, significantly greater integration of the pathogens into pre-established commensal populations occurred, resulting in populations of the pathogens that reached similar densities to the control substrata. This may result from detachment of the pre-established bacterial communities in response to rapidly increased nutrient availability as the growth medium was substituted, possibly allowing greater adherence of the pathogenic species. This is supported by the observation that the resistance to colonisation was greater when S. saprophyticus and C. xerosis were continually grown in simulated serum.
The intention of this investigation was not to directly simulate the epidermal and wound environment, but rather, to assess the potential of a biofilm model utilising a novel fine-celled foam substratum for use in investigations into physiological and ecological interactions between specific groups of wound and skin isolates, under controlled conditions. Colonisation resistance, which varied depending on the medium environment and the commensal and pathogenic bacteria, could be broadly quantified in the FCF model. The model could be further applied to investigate bacteriotherapy for wounds and to model the effects of selective antimicrobial treatments on the bacteriological composition of biofilms grown using wound isolates.
Disclosure statement
No potential conflict of interest was reported by the authors.
Funding
Part of this work was done during a studentship that was supported by grants from the BBSRC [grant number BB/D526588/1] and ConvaTec.
References
- Agostinho AM, Hartman A, Lipp C, Parker AE, Stewart PS, James GA. 2011. An in vitro model for the growth and analysis of chronic wound MRSA biofilms. J Appl Microbiol. 111:1275–1282. doi:10.1111/jam.2011.111.issue-5
- Al-Bakri AG, Gilbert P, Allison DG. 2004. Immigration and emigration of Burkholderia cepacia and Pseudomonas aeruginosa between and within mixed biofilm communities. J Appl Microbiol. 96:455–463. doi:10.1111/jam.2004.96.issue-3.
- Axelsson L, Mahida Y. 2000. Flora: role in colonisation resistance and other effects; production of antimicrobial peptides. Microb Ecol Health Dis. 12;1 supp 2:216 –422.
- Barnea Y, Weiss J, Gur E. 2010. A review of the applications of the hydrofiber dressing with silver (Aquacel Ag(®)) in wound care. Ther Clin Risk Manag. 6:21–27.
- Bjarnsholt T, Kirketerp-Moller K, Jensen PO, Madsen KG, Phipps R, Krogfelt K, Hoiby N, Givskov M. 2008. Why chronic wounds will not heal: a novel hypothesis. Wound Repair Regen. 16:2–10. doi:10.1111/wrr.2008.16.issue-1.
- Charles CA, Ricotti CA, Davis SC, Mertz PM, Kirsner RS. 2009. Use of tissue-engineered skin to study in vitro biofilm development. Dermatol Surg. 35:1334–1341. doi:10.1111/j.1524-4725.2009.01238.x.
- Dalton T, Dowd SE, Wolcott RD, Sun Y, Watters C, Griswold JA, Rumbaugh KP. 2011. An in vivo polymicrobial biofilm wound infection model to study interspecies interactions. PLoS one. 6:e27317. doi:10.1371/journal.pone.0027317.
- Dowd S, Sun Y, Secor P, Rhoads D, Wolcott B, James G, Wolcott R. 2008. Survey of bacterial diversity in chronic wounds using pyrosequencing, DGGE, and full ribosome shotgun sequencing. BMC Microbiol. 8:43. doi:10.1186/1471-2180-8-43.10.1186/1471-2180-8-43
- Dowd SE, Sun Y, Smith E, Kennedy JP, Jones CE, Wolcott R. 2009. Effects of biofilm treatments on the multi-species Lubbock chronic wound biofilm model. J Wound Care. 18(508):510–512.
- Fredricks DN. 2001. Microbial ecology of human skin in health and disease. J Investig Dermatol Symp Proc. 6:167–169. doi:10.1046/j.0022-202x.2001.00039.x.
- Gander S, Hayward K, Finch R. 2002. An investigation of the antimicrobial effects of linezolid on bacterial biofilms utilizing an in vitro pharmacokinetic model. J. Antimicrob. Chemother. 49:301–308.
- Haisma EM, Rietveld MH, de Breij A, van Dissel JT, El Ghalbzouri A, Nibbering PH. 2013. Inflammatory and antimicrobial responses to methicillin-resistant staphylococcus aureus in an in vitro wound infection model. PLoS one. 8:e82800. doi:10.1371/journal.pone.0082800.
- Hammond AA, Miller KG, Kruczek CJ, Dertien J, Colmer-Hamood JA, Griswold JA, Horswill AR, Hamood AN. 2011. An in vitro biofilm model to examine the effect of antibiotic ointments on biofilms produced by burn wound bacterial isolates. Burns. 37:312–321. doi:10.1016/j.burns.2010.09.017.
- Hill KE, Malic S, McKee R, Rennison T, Harding KG, Williams DW, Thomas DW. 2010. An in vitro model of chronic wound biofilms to test wound dressings and assess antimicrobial susceptibilities. J Antimicrob Chemother. 65:1195–1206. doi:10.1093/jac/dkq105.
- Humphreys G, Lee GL, Percival SL, McBain AJ. 2011. Combinatorial activities of ionic silver and sodium hexametaphosphate against microorganisms associated with chronic wounds. J Antimicrob Chemother. 66:2556–2561. doi:10.1093/jac/dkr350.
- Inniss WE, Mayfield CI. 1978. Growth rates of psychrotrophic sediment microorganisms. Water Res. 12(4):231–236. doi:10.1016/0043-1354(78)90091-X.
- James G, Swogger E, Wolcott R, deLancey-Pulcini E, Secor P, Sestrich J, Costerton JW, Stewart PS. 2008. Biofilms in chronic wounds. Wound Repair Regen. 16:37–44. doi:10.1111/wrr.2008.16.issue-1.
- Kinniment SL, Wimpenny JWT, Adams D, Marsh PD. 1996. Development of a steady-state oral microbial biofilm community using the constant-depth film fermenter. Microbiology. 142:631–638. doi:10.1099/13500872-142-3-631.
- Kostenko V, Lyczak J, Turner K, Martinuzzi RJ. 2010. Impact of silver-containing wound dressings on bacterial biofilm viability and susceptibility to antibiotics during prolonged treatment. Antimicrob Agents Chemother. 54:5120–5131. doi:10.1128/AAC.00825-10.
- Kucera J, Sojka M, Pavlik V, Szuszkiewicz K, Velebny V, Klein P. 2014. Multispecies biofilm in an artificial wound bed—A novel model for in vitro assessment of solid antimicrobial dressings. J Microbiol Meth. 103:18–24. doi:10.1016/j.mimet.2014.05.008.
- Ledder RG, Madhwani T, Sreenivasan PK, De Vizio W, McBain AJ. 2009. An in vitro evaluation of hydrolytic enzymes as dental plaque control agents. J Med Microbiol. 58:482–491. doi:10.1099/jmm.0.006601-0.
- Lenski RE, Rose MR, Simpson SC, Tadler SC. 1991. Long-term experimental evolution in Escherichia coli. 1. Adaptation and divergence during 2,000 generations. Am Soc Nat. 138:1315–1341. doi:10.1086/an.1991.138.issue-6.
- Lipp C, Kirker K, Agostinho A, James G, Stewart P. 2010. Testing wound dressings using an in vitro wound model. J Wound Care. 19:220–226. doi:10.12968/jowc.2010.19.6.48468.
- Malic S, Hill KE, Hayes A, Percival SL, Thomas DW, Williams DW. 2009. Detection and identification of specific bacteria in wound biofilms using peptide nucleic acid fluorescent in situ hybridization (PNA FISH). Microbiology. 155:2603–2611. doi:10.1099/mic.0.028712-0.
- Matejka KM, Bremer PJ, Tompkins GR, Brooks HJL. 2012. Antibiotic susceptibility of Moraxella catarrhalis biofilms in a continuous flow model. Diagn Microbiol Infect Dis. 74:394–398. doi:10.1016/j.diagmicrobio.2012.08.021.
- McAuliffe O, Ross RP, Hill C. 2001. Lantibiotics: structure, biosynthesis and mode of action. Fems Microbiol Rev. 25:285–308. doi:10.1111/j.1574-6976.2001.tb00579.x.
- McBain AJ. 2009. Chapter 4 In vitro biofilm models: an overview. Adv Appl Microbiol. 69:99–132. doi:10.1016/S0065-2164(09)69004-3.
- McBain AJ, Bartolo RG, Catrenich CE, Charbonneau D, Ledder RG, Gilbert P. 2003a. Growth and molecular characterization of dental plaque microcosms. J Appl Microbiol. 94:655–664. doi:10.1046/j.1365-2672.2003.01876.x.
- McBain AJ, Bartolo RG, Catrenich CE, Charbonneau D, Ledder RG, Rickard AH, Symmons SA, Gilbert P. 2003. Microbial characterization of biofilms in domestic drains and the establishment of stable biofilm microcosms. Appl Environ Microbiol. 69:177–185. doi:10.1128/AEM.69.1.177-185.2003.
- McBain AJ, Ledder RG, Moore LE, Catrenich CE, Gilbert P. 2004. Effects of quaternary-ammonium-based formulations on bacterial community dynamics and antimicrobial susceptibility. Appl Environ Microbiol. 70:3449–3456. doi:10.1128/AEM.70.6.3449-3456.2004.
- Oates A, Bowling FL, Boulton AJM, Bowler PG, Metcalf DG, McBain AJ. 2014. The visualization of biofilms in chronic diabetic foot wounds using routine diagnostic microscopy methods. J Diabetes Res. 2014:8. doi:10.1155/2014/153586.
- Pratten J, Smith AW, Wilson M. 1998. Response of single species biofilms and microcosm dental plaques to pulsing with chlorhexidine. J Antimicrob Chemother. 42:453–459. doi:10.1093/jac/42.4.453.
- Rozen DE, McGee L, Levin BR, Klugman KP. 2007. Fitness costs of fluoroquinolone resistance in streptococcus pneumoniae. Antimicrob Agents Chemother. 51:412–416. doi:10.1128/AAC.01161-06.
- Russell R, Wiles CM. 1970. Documenta Geigy Scientific Tables. 7th ed. Ciba-Geigy Ltd.
- Steffansen B, Herping SPK. 2008. Novel wound models for characterizing ibuprofen release from foam dressings. Int J Pharm. 364:150–155. doi:10.1016/j.ijpharm.2008.07.025.
- Sun Y, Dowd SE, Smith E, Rhoads DD, Wolcott RD. 2008. In vitro multispecies Lubbock chronic wound biofilm model. Wound Repair Regen. 16:805–813. doi:10.1111/j.1524-475X.2008.00434.x.
- Taylor B, Greenman J. 2010. Modelling the effects of pH on tongue biofilm using a sorbarod biofilm perfusion system. J Breath Res. 4:017107. doi:10.1088/1752-7155/4/1/017107.
- Thorn RMS, Greenman J. 2009. A novel in vitro flat-bed perfusion biofilm model for determining the potential antimicrobial efficacy of topical wound treatments. J Appl Microbiol. 107:2070–2079. doi:10.1111/jam.2009.107.issue-6.
- Trengove, Langton. 1996. Biochemical analysis of wound fluid from nonhealing and healing chronic leg ulcers. Wound Repair Regen. 4:234–239. doi:10.1046/j.1524-475X.1996.40211.x.
- Varella Coelho ML, Santos Nascimento JD, Fagundes PC, Madureira DJ, Oliveira SSD, de Paiva Vasconcelos, Brito MA, Freire Bastos MDCD. 2007. Activity of staphylococcal bacteriocins against Staphylococcus aureus and Streptococcus agalactiae involved in bovine mastitis. Res Microbiol. 158:625–630. doi:10.1016/j.resmic.2007.07.002.
- Woods J, Boegli L, Kirker KR, Agostinho AM, Durch AM, deLancey Pulcini E, Stewart PS, James GA. 2012. Development and application of a polymicrobial, in vitro, wound biofilm model. J Appl Microbiol. 112:998–1006. doi:10.1111/jam.2012.112.issue-5.
- Wright E, Neethirajan S, Weng X. 2015. Microfluidic wound model for studying the behaviors of Pseudomonas aeruginosa in polymicrobial biofilms. Biotechnol Bioeng. 112:2351–2359. doi:10.1002/bit.v112.11.
- Yang Q, Phillips PL, Sampson EM, Progulske-Fox A, Jin S, Antonelli P, Schultz GS. 2013. Development of a novel ex vivo porcine skin explant model for the assessment of mature bacterial biofilms. Wound Repair Regen. 21:704–714. doi:10.1111/wrr.12074.