Abstract
Cooling and lubrication agents like triethanolamine (TEA) are essential for many purposes in industry. Due to biodegradation, they need continuous replacement, and byproducts of degradation may be toxic. This study investigates an industrial (1,200 m³) cooling-lubrication circuit (CLC) that has been in operation for 20 years and is supposedly in an ecological equilibrium, thus offering a unique habitat. Next-generation (Illumina Miseq 16S rRNA amplicon) sequencing was used to profile the CLC-based microbiota and relate it to TEA and bicine dynamics at the sampling sites, influent, machine rooms, biofilms and effluent. Pseudomonas pseudoalcaligenes dominated the effluent and influent sites, while Alcaligenes faecalis dominated biofilms, and both species were identified as the major TEA degrading bacteria. It was shown that a 15 min heat treatment at 50°C was able to slow down the growth of both species, a promising option to control TEA degradation at large scale.
Introduction
Triethanolamine (TEA; C6H15NO3) is an alkanolamine that is frequently used as a main chemical working component in industrial manufacturing plants (Speranza et al. Citation2006). Its chemical properties makes it a popular cutting and metal working fluid as well as corrosion inhibitor (Weavers et al. Citation1997). Furthermore, it is used in industrial, pharmaceutical and cosmetic products as emulsifier and pH buffer (Sandin et al. Citation1990; Lessmann et al. Citation2009; Libralato et al. Citation2010). Alkanolamines are highly susceptible to biodegradation in various environments like soil and fresh water. Therefore, bacterial deterioration of TEA in water-soluble working fluids is a well-known problem (Prince et al. Citation1994; Eide-Haugmo et al. Citation2009). With concentrations around 1% (w v–1), TEA represents the major carbon and nitrogen source in many cooling liquid circuits (CLC) of various production sites, leading to selective enrichment and accumulation of TEA degrading bacteria. Although biodegradability can be seen as advantageous for the disposal after use (Buers et al. Citation1997), TEA degraders need to be identified and studied in more detail in order to control deterioration processes and bacterial colonization during its industrial application. In particular, bicine (C6H13NO4) is considered an undesired degradation product that may accelerate equipment corrosion.
Degradation of working fluids like TEA causes additional expense due to the need for continuous replenishment to ensure proper process performance. The hydraulic retention time of a cooling liquid often adds up to several months and its replacement is an environmental and economic burden. A high retention time favors accumulation of microorganisms, both in the cooling liquid as well as in biofilms. Mechanical solutions like chain-driven collectors or rakes able to remove solid particles, including microbial aggregates and biofilms, are usually the sole means of suppressing microbial growth. Due to the inefficiency of such measures, biocides are applied, offsetting the advantages of environmentally friendly lubricants like TEA. To counteract the development of microbial resistance, new biocides have to be implemented every few months (Mattsby-Baltzer et al. Citation1989; Simões et al. Citation2011). Still, biofilms may continue to become established in pipes and basins, and microbial colonizers and degradation products may continue to pose safety hazards for workers exposed to the cooling liquid (Grant and Bott Citation2005; Lessmann et al. Citation2009). Biofilms, in addition, may not only contribute to the corrosion of pipes, basins and machinery, but also serve as a continuous source of TEA degraders (Adapa et al. Citation2016).
Despite their low toxicity at neutral pH, alkanolamines are known for their toxicity towards microorganisms under alkaline conditions, inhibiting microbial growth at pH 9 (Sandin et al. Citation1990; Bakalova et al. Citation2014). Alkalinity in production circles, however, can pose health risks for workers and can negatively impact the equipment.
In this study, four sections within an industrial CLC were analyzed (influent, effluent, biofilm and machines). The aim was to investigate the microbial community composition in an industrial CLC both from planktonic and biofilm sources and to examine possible ways of inhibiting the growth of microorganisms capable of TEA degradation. It was hypothesized (1) that the microbiota at the four different sampling points would deviate from each other and (2) that TEA degradation could be inhibited by a short-term thermal treatment within the circuit or (3) by increasing the pH to 9.
Material and methods
Sampling
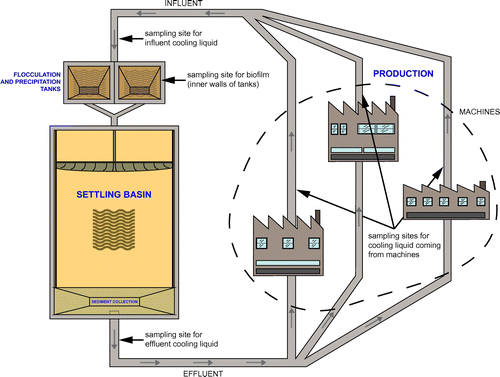
Within the CLC (total volume of 1,200 m3, various basins, 1,700 m piping; Figure ) four sampling sites were chosen for their different exposure to the environment and by the possible influence of structural site-specific characteristics on the microbiota: (1) biofilm built up in flocculation tanks above the water line and constantly humidified by turbulence, (2) the effluent cooling liquid after passage through the settling basins, (3) the cooling liquid taken directly from machines and (4) the influent cooling liquid heading from machines to flocculation tanks. The sampling took place on Mondays and Fridays over the course of three weeks in November 2015. The liquid samples (V = 1 l each) were taken with a scooper, and biofilm samples (30 g fresh weight) were scraped from the inner walls of the precipitation tanks and placed in sterile glass test tubes. Effluent, influent and biofilm samples were collected on each of the six sampling days. Machine samples were taken on Mondays after work-free weekends when the main circuit was bypassed and the circuit liquid did not run through the machines. The samples from three machines were taken on the first Monday only, and samples from three machine rooms on the second and third Monday (making up a total of nine machine samples).
Physico-chemical parameters
Temperature, oxygen concentrations and pH were measured daily by online measurement probes installed on the industrial site (for pH measurement: Orbisint CPS11D-7BA21 Memosens by Endress+Hauser, Reinach BL, Switzerland; for O2 measurement: TriOxmatic® 700 IQ by WTW, Weilheim, Germany; for temperature measurement: EasyTemp™ TSM470-B1D11FC and TMP31-A1XABBAE1AAA both by Endress+Hauser, Reinach BL, Switzerland). TEA and bicine concentrations were determined weekly by Thermo Scientific®, Dionex™ ICS-1100 ion chromatograph using a Dionex™ Autosampler AS-40 and a Shodex refraction index detector RI-71 using internal standards of triethanolamine and bicine ranging from 0 mg l−1 to 500 mg l−1. Data were processed with the Dionex™ Chromaeleon Data System (all Thermo-Fischer Scientific, Vienna, Austria).
DNA extraction
To gather the biomass for the DNA extraction, 400 ml of the effluent samples and 200 ml of the influent and machine samples were filtered via a vacuum filtration system (Merck Millipore™, Merck, Darmstadt, Germany) using UV-sterilized 0.2 μm cellulose filters (GE Healthcare Life Sciences Whatman™ RC 58 Membrane Filters, VWR, Vienna, Austria). The filters with the filter cake were cut into halves, and each was then placed in 1.5 ml microcentrifuge tubes and frozen until DNA extraction.
Prior to DNA extraction 900 μl of buffer (50 mM glucose, 25 mM tris(hydroxymethyl)aminomethane (TRIS), 10 mM thylenediaminetetraacetic (EDTA), pH 8) (both Merck, Darmstadt, Germany), and 150 μl of 0.5 M EDTA (pH 8) (Applichem, Graz, Austria) were added to each microcentrifuge tube containing half a filter. The tubes were vortexed thoroughly to dissociate the pellet from the filter. Finally, filters were rinsed with buffer to assure that all cells were detached from the filter and the tube contents of two half filters were pooled. For biofilm samples the same buffer and EDTA solutions were used per tube containing 1 g of biofilm material. DNA was extracted using the NucleoSpin® Soil-Kit (Macherey-Nagel Citation2014/Rev. 04, Düren, Germany) following the user manual. For the extraction process, tubes with the pooled contents of both filter halves of the liquid samples and the tubes containing the biofilm samples were centrifuged at 10,000 g for 10 min. The pellets were used as starting material for the kit-protocol.
Next generation sequencing
DNA extracts from all samplings were prepared using the Macherey-Nagel DNA extraction kit according to the manufacturer’s protocol and contained 26.8 ± 11.4 ng μl−1 of DNA. Sequencing of the samples was carried out at Microsynth AG (Balgach, Switzerland) on the Illumina MiSeq platform using the 2 × 250 base paired-end approach according to the company’s standard protocol. Briefly, the processing through Microsynth AG included preparation of the Nextera XT library, demultiplexing, removal of Illumina adaptors and locus specific primers and stitching of the trimmed reads. The primers used were 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) (V4).
Cultivation and growth experiments
For quantifying and isolating bacteria, a cooling-liquid-medium (CLM) was prepared by autoclaving and subsequent filtering (0.2 μm Whatman® RC 58 Cellulose Membrane Filters; Merck, Darmstadt, Germany) of cooling liquid. Autoclaving sterilized the cooling liquid and inactivated the heat-labile biocide (methylisothiazolinone). Filtering removed precipitates and other coarse solid particles. For isolation of TEA degraders, a synthetic TEA growth medium was prepared (Knapp et al. Citation1996) and adjusted to pH 7.0. Autoclaving did not modify the chemical composition with respect to TEA and bicine, as confirmed by ion chromatography. However, the heat inactivated the thermo-labile biocide. To estimate the number of colony forming units (CFU), the samples were serially diluted, plated in triplicates on agar (15 g l−1 CLM) plates and incubated for a maximum time of 144 h.
From the plate-grown populations, 41 colonies were randomly picked with sterile pipette tips for all sampling sites except machines and transferred to fresh CLM and TEA plates. For each site, 10 random colonies that manifested growth on both CLM and TEA plates were picked and transferred to fresh CLM master plates. The cultivation on TEA agar acted as verification for the ability of the isolates to use TEA as sole carbon source. All plates were incubated at 32°C, which compares to the mean temperature in the circuit.
The inoculum for the growth tests was taken from well-grown liquid cultures (OD600 of ~1.5). The biofilm isolates were also grown in liquid culture. Growth experiments in liquid culture were carried out in triplicates in 100 ml conical flasks with 50 ml of CLM + 2 ml of inoculum at 32°C using a Multitron Standard incubator shaker (Infors AG, Bottmingen-Basel, Switzerland; 120 rpm). Microbial growth was quantified by optical density (OD600) at different time points using a spectrophotometer (Hitachi U-2001 UV/Vis Spectrophotometer). During the exponential stage, samples for TEA and bicine quantification were taken and stored for a maximum of four days at 4°C until analysis. The sampling continued until the stationary phase was reached.
Bacterial growth rates were calculated from the change in OD600 h−1 during the exponential growth phase as follows:
For the thermal inhibition experiment, the five isolates showing the highest growth rates in suspended cultures in TEA medium were selected (they were all from biofilm samples) and temperature-treated for 5 and 15 min at 50°C, as well as cultured at pH 9 (adjusted with 5 M NaOH). Again, triplicates of 50 ml of CLM were inoculated with 2 ml of preculture (OD600 at around 1.5) and the growth conditions in the liquid culture were set as explained above. Replicates where growth showed retardation were measured for 100 h. Reactions without any sign of growth inhibition were terminated after 50 h.
Measurement of TEA and bicine concentrations
To depict the degradation of TEA and increase in bicine, concentrations at the beginning of the growth experiments as well as during and at the end of exponential growth phase were measured using ion chromatography. Samples were diluted in 0.088 M KH2PO4 (pH 2.6) and measured in triplicates. TEA and bicine solutions (0–500 mg l−1) served as standards (R² = 0.9998). As the growth experiments were conducted in three batches (each site separately), initial concentrations (min0) of both TEA and bicine varied between cycles due to fluctuating TEA and bicine concentrations in the circuit and therefore in the prepared medium.
Sequencing of isolates
Bacterial isolates were identified by 16S Sanger sequencing from pure culture DNA extracts. A cell pellet from 1 ml of culture was used as starting material for DNA extraction as described above. Amplification was conducted in volumes of 25 μl each, consisting of 1 μl of DNA-extract, 50 μM dNTP each, 50 μM each primer (8f: 5′-AGAGTTTGATCCTGGCTCAG-3′, 1492r: 5′-TACGGYTACCTTGTTACGACTT-3′), 25 μM MgCl2, 10 μg of BSA, 1-fold reaction buffer and 0.625 U MyTaq polymerase following the PCR protocol of Loy et al. (Citation2002). PCR products were purified using the Sigma Aldrich® GenElute™ PCR Clean-Up kit to remove primer residues, unused dNTPs and enzymes. DNA was quantified using a NanoDrop™ 2000c spectrophotometer (Thermo Fisher Scientific Inc., Waltham, USA). Sequencing was carried out by Eurofins Genomics using the primers used for amplification (8f and 1492r).
Bioinformatics
Sequencing data were analyzed in mothur v.1.37.4 (Schloss et al. Citation2009) following the MiSeq SOP (Kozich et al. Citation2013). PCR and sequencing errors were reduced by filtering sequence reads for length, ambiguity and homopolymorphism. Chimeric sequences were removed using uchime algorithm integrated in mothur (Edgar et al. Citation2011). For the alignment, the SILVA bacteria database (www.arb-silva.de)(Yilmaz et al. Citation2014) served as reference and RDP v14 reference files were used for the taxonomic classification (cutoff = 60). Prior to clustering of the sequences at 97% similarity level, reads belonging to the unspecific lineages of chloroplasts, mitochondria, unknown sequences and Eukaryota were removed. Rare operational taxonomic units (OTUs) with a total abundance of two reads were removed. Groups were subsampled according to the smallest sample size. Calculation of Venn diagrams was used to assess numbers of shared and unique OTUs in samples.
Multiple sequence alignment of sequenced isolates was conducted in R (v.3.3.1) (R Development Core Team Citation2008) using the msa package (Bodenhofer et al. Citation2015) and ClustalW algorithm (Thompson et al. Citation1994). Assignment of sequencing results to the nucleotide collection database was carried out through nucleotide BLAST® search.
Statistical analysis
Non-metrical multidimensional scaling (NMDS) was used to compress the multidimensional character of the OTU table resulting from bioinformatic analysis into two dimensions. Ordination was calculated on relative abundance data in R v.3.3.1 (R Development Core Team Citation2008) using metaMDS function of the vegan package (Oksanen et al. Citation2016). Linear discriminant analysis of effect size (LEfSe was executed in mothur v.1.37.4 as integrated function; Segata et al. Citation2011) was performed on OTU relative abundance data to assess distances between sampling sites and highlight taxa that are accountable for differences in community composition of examined sites. Taxonomic units and phylogenetic groups with LDA ≥ 2 and a p-value < 0.05 were considered significant. Graphics were designed in R using ggplot2 (Wickham Citation2009). To assess significant changes in OTU abundances over time and between sites, analysis of molecular variance (AMOVA) was performed in mothur v.1.37.4 with OTU data grouped by sampling-week and sampling-site, respectively. Differences were significant in a 95% confidence interval. To analyze the dissimilarity matrix and illustrate minimum, mean and maximum dissimilarity, disana (dissimilarity analysis) was conducted in R using the labdsv package (Roberts Citation2016). Correlations between variables were calculated as Pearson’s r or Spearman’s rho correlation coefficient.
Results and discussion
The investigated CLC which has been in continuous operation since 1998 represents a particular industrial ecosystem which supposedly is in an equilibrium. The uniqueness of the study is that such system, and various subsections thereof, has never been analyzed by an NGS approach targeting the microbiota despite obvious effects of its colonization on production performance, such as TEA degradation. On the basis of isolates identified as TEA degraders, the final goal was to find a solution for slowing down the degradation of TEA. The first aim was to estimate bacterial numbers in the CLC of the industrial manufacturing plant at different sites. CLM plates biofilm (8.0 × 105 to 2.0 × 106 CFU ml−1) and influent samples (3.1 × 105 to 1.34 × 106 CFU ml−1) showed the highest cell count, whereas low numbers were observed in the effluent (between 5.7 × 104 and 1.6 × 105 CFU ml−1) and machine derived samples (no growth) (Figure A). The same pattern, with slightly lower counts, was found on TEA agar which is more selective for TEA degraders than CLM. During the four days of CLM and TEA plate incubation, the plates were checked daily. Upon visual examination, bacterial growth was observed on CLM agar before colonies on TEA agar reached a detectable size. Consequently, growth was not only faster on CLM plates, but colonies were also larger compared to the TEA agar. Colony morphology showed little diversity and was mostly limited to round pale yellow colonies on CLM agar and small white colonies on TEA agar. CFU numbers in influent samples were on average 80% higher than in effluent samples indicating the considerable efficiency of the sedimentation basin. To assure proper functioning of the CLC, temperature, pH, O2 and concentrations of biocide, TEA and bicine in the CLC were continuously monitored by plant operators. Since the physico-chemical parameters of the cooling liquid were expected to be balanced throughout the larger part of the circuit, probes for monitoring were located only in the settling basin. This provides better accessibility and facilitates handling but neglects possible site-specific fluctuations. Major differences between sites occur mostly with regards to O2 concentration caused by microbial oxygen consumption and lack of O2 input in closed piping systems. Generally, however, O2 fluctuations in the circuit can be inferred from the monitoring in the settling basins (Figure ).
Figure 2. Bacterial abundance and community composition. (A) Colony forming units on CLM and TEA agar of the effluent, influent and biofilm samples. Results from machine samples are not shown, since no growth was observed. (B) Overview of community composition on class level in four sampled sites with discrimination between weekdays. Friday and Monday samples were pooled for each site, as there was no community shift in the course of the three-week sampling period. Groups accounting for <1% are grouped in Other. (BF = biofilm Fridays, BM = biofilm Mondays, EF = effluent Fridays, EM = effluent Mondays, IF = influent Fridays, IM = influent Mondays, MM = machines Mondays). (C) Non-metric multidimensional scaling using OTUs and sites. (D) Excerpt of results from linear discriminant analysis of effect size (LEfSe) based on OTU data and representing significantly overrepresented genera with highest LDAs at each sample location.
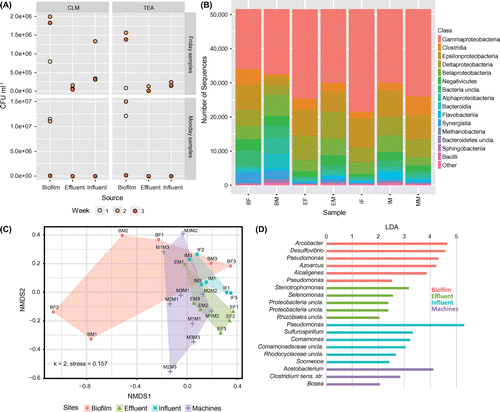
Figure 3. Oxygen concentrations and pH measured via probes in the settling basin during the sampling period. Dotted vertical lines represent each of the six sampling time-points.
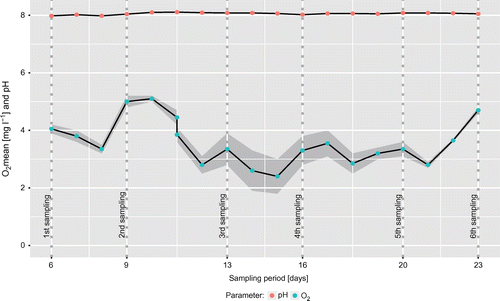
The pH remained stable at 8.06 ± 0.04 and had no measurable effect on bacterial growth. Biocide levels averaged at 337 ± 29 mg l−1 and were not correlated to microbial growth and oxygen concentrations. Oxygen concentrations ranging between 5.15 and 2.40 mg O2 l−1 mostly decreased during the week and recovered over the weekend (Figure ). The CFU was negatively associated with the oxygen concentration (meanPearson’s r across sample groups = –0.31), which suggests an elevated microbial activity during workdays compared to work-free days, probably due to nutrient or substrate input from the production, or a disruption of established microbial aggregates. This is underlined by a higher bacterial count at the end of the week (Figure A). In contrast to the other sites, where no growth was observed on Monday samples, CFU numbers in biofilm were similar for Monday and Friday samples (7.5 × 106 ± 5.2 × 106 and 1.5 × 106 ± 0.53 × 106, respectively).
A strong negative correlation (r = –0.97) between TEA (7.85–6.57 mg l−1) and bicine concentration (1.16–1.76 mg l−1) was found. In the CLC, bicine concentrations were monitored to serve as a proxy for cooling liquid decay and bacterial contamination since plant operators observed a correlation between bacterial abundance and bicine concentration. Several mechanisms for bicine formation have been proposed before, mainly in connection with gas treatment (Fytianos et al. Citation2016). Oxidation of TEA or the influence of heat on methyldiethanolamine or diethanolamine can accelerate bicine formation (Lawson et al. Citation2003). In the CLC, bicine concentrations showed a delayed rise compared to decreases in TEA. The pathways of microbial TEA degradation under anaerobic conditions were already published (Frings et al. Citation1994; Knapp et al. Citation1996; Speranza et al. Citation2006), but no information on bacterial bicine production from TEA is available in the literature. If bicine was solely produced by TEA oxidation, a positive correlation would be expected between O2 and bicine concentrations. However, this correlation was weak and not significant, indicating other sources of bicine production than TEA oxidation. Based on the above-mentioned observations in earlier work and on the data from the present study, a connection between bacterial bicine production and TEA degradation is likely.
Bacterial abundance and community composition
A detailed insight into the community composition at different sampling sites was provided by Illumina Miseq sequencing. A total number of 1.4 × 106 subsampled and processed 16S targeted reads was generated (1.6 × 105 ± 4.5 × 104 reads per sample) which were clustered into 1,477 OTUs, of which 471 could not be found across all sampling sites. Rarefaction curves nearly reached saturation, indicating that sequencing depth was sufficient for a representative picture of the bacterial community. Dissimilarity analysis (Disana) exhibited a high average dissimilarity (Bray–Curtis measure) of 0.3 between most samples.
The number of observed OTUs (1,020) was highest in machine samples, which is in line with the hypothesis that the production process supplements the circuit with nutrients. The lowest richness was observed in effluent, with a total of 796 OTUs, probably due to elimination in the settling basin. The highest number of shared OTUs was found for machine and influent samples, which is coherent with the course of the cooling liquid circuit (Figure ).
Gammaprotebacteria appeared as the most dominant class (>23% in each sample) in all samples, with Pseudomonas being the most prevalent representative of this group (Figure B). Obligate anaerobic clostridia also accounted for a considerable fraction of sequences in all samples, including the biofilm. Biofilms are known to become anaerobic within a few millimeters and pipes in the circuit are up to 1,700 m in length, during which no O2 input occurs. Oxygen is depleted by the high numbers of aerobic species, which in turn can facilitate propagation of anaerobic microbes in the passageways of the circuit that are not exposed to air. The degradation of TEA is a known capability of several anaerobic organisms, including members of the class Clostridia (Frings et al. Citation1994; Speranza et al. Citation2006), which also increased in abundance in biofilm samples (Figure B). In machine samples clostridial abundances were twice as high (>12%) as in other sites (< 6%). Their known capacity to exploit TEA via anaerobic degradation using ethanolamine ammonia lyases (Frings et al. Citation1994) suggests an active clostridial contribution to TEA degradation in industrial CLCs. In this study, specific microbial activities have not been taken into consideration. As an estimate, the ratio of intra-/extra-cellular DNA might be used (Nagler et al. Citation2018), as DNA is considered to be a major component of the extracellular polymeric substances (EPS) in biofilms (Flemming and Wingender Citation2010). This information might strengthen the allocation of main TEA degradation to the biofilms in the CLC and should be considered in future experiments.
In addition to obvious phylogenetic differences in bacterial communities among sites (Figure B), smaller scale compositional differences can also be seen from the distances between polygons in Figure C (NMDS). The overlapping areas represent related community structures between sites, while site-specific circuit features account for differences between communities. The area of depicted polygons is defined by group heterogeneity. In line with richness on the sites, the heterogeneity within a group was largest in biofilm and smallest in influent samples. Underlining a stable bacterial community and representative sampling, communities from the same site were similar, irrespective of sampling time (AMOVA p = 0.22, Figure C). However, there were significant differences in bacterial community composition among the sites (p = 0.003). Biofilm samples differed from effluent (p = 0.026) and influent samples (p = 0.035) and machine samples differed from influent samples (p = 0.016). No significant differences between influent-effluent and biofilm-machines were detected (p = 0.080 and p = 0.057, respectively). Despite the sedimentation in the settling basin, effluent and influent samples were similar in their community composition, which suggests that the settling process is not selective. The difference between biofilm and influent-effluent samples indicates that the biofilm microbiota is not solely a sedimentation of the liquid CLC system but offered and created conditions for a specific community composition, as has been shown for other habitats (Toyofuku et al. Citation2015; Liu et al. Citation2009). The fact that biofilm is not constantly submerged in cooling liquid and therefore not in steady contact with the circuit flow might have supported the development of a different community composition (Figure C).
From LefSe analysis (Segata et al. Citation2011; Figure D), 53 OTUs were found significantly overrepresented at a certain site, with most of them belonging to Beta- (13 OTUs), Gamma- (10 OTUs) and Alphaproteobacteria (7 OTUs). Clostridia, which were abundant in all samples, only accounted for 5 OTUs characteristic for a specific site. In the biofilm, OTUs classified as Pseudomonas sp., Arcobacter sp., Desulfovibrio sp., Azoarcus sp. and Acaligenes sp. were found to be significantly overrepresented and may therefore be regarded as characteristic members of the biofilm microbiota. All of them are known for biofilm formation and can be found in contaminated watery environments (Teixeira and Oliveira Citation2002; Benedek et al. Citation2016; Auguet et al. Citation2017; Jia et al. Citation2018). While Pseudomonas was the most abundant genus in all sites, specific Pseudomonas OTUs appeared to be distinctive for influent (linear discriminant analysis score, LDA 5.3) and biofilm samples (LDA 4.3 and 2.5). From an overall of 19 Pseudomonas OTUs, two were found in all biofilm samples; one of the two OTUs was also characteristic for the influent. One Pseudomonas OTU was only found in the biofilm samples (Figure ), and one was found both in biofilm and influent, while 17 were spread over all sites. The long-term survival of Pseudomonas sp. under oxygen-depleted conditions without transitioning in a dormant state using extracellular electron shuttles for fermentation may have contributed to its dissemination (Glasser et al. Citation2014). These findings suggest that the sought-after aerobic TEA degraders are representatives of these Pseudomonas OTUs.
The high abundance of aerobic bacteria in the effluent may be explained by the oxygen input from the upstream flocculation and aeration process in the circuit. Subsequently, while heading to the machines, O2 in the cooling liquid is exhausted and anaerobicity increased. Anaerobic bacteria, among them clostridia, were detected in all sections of the circuit, probably due to carryover and their ability to form endospores. The flow rate is not consistent throughout the system; therefore, due to homogeneous dispersion, spores could accumulate in dead zones of the circuit. The ability of rapid reactivation from dormancy may allow for site-specific activities as soon as anaerobic conditions are given (Sinai et al. Citation2015).
Machine samples were defined by the presence of strictly anaerobic Acetobacterium (LDA 4.1), a member of the class Clostridia, and Clostridium sensu stricto, which supports the differences in bacterial community composition, especially the increase in anaerobic species (Figure B). Oxygen input in the machine area during the weekends is negligible, thus providing beneficial conditions for anaerobic bacteria which might also support biofilm formation. Further analyses involving biofilm formation rates and degradation tests under anaerobic or microaerophilic conditions will be necessary to evaluate the contribution of specific anaerobic community members like Acetobacterium sp. and Clostridium sp. to TEA degradation. Typical for the poorly colonized effluent samples was the aerobic Stenotrophomonas (LDA 3.2), which, like Pseudomonas, is a member of the Gammaproteobacteria.
Growth experiments without treatment
The connection between TEA degradation and bicine production was investigated in growth experiments with 30 bacterial isolates able to grow on TEA as sole carbon source. All sequenced isolates showed affiliation to two highly abundant genera which were distinctive for biofilm samples (Figure D). All isolates originating from effluent and influent samples were identified as Pseudomonas pseudoalcaligenes (identity ≥ 99%) while six out of 10 biofilm isolates were found to be Alcaligenes faecalis (identity ≥ 99%), with the other four being P. pseudoalcaligenes (Table ). Pseudomonas spp., especially P. pseudoalcaligenes, are widespread in industrial working fluids (Mattsby-Baltzer I et al. Citation1989; Sandin et al. Citation1990). A. faecalis is ubiquitously found in water and soil (Bizet and Bizet Citation1997), but has not previously been found in cooling circuits.
Table 1. Identification of randomly selected isolates from the four sampling sites.
Growth rates were calculated from OD600 change during the exponential phase. Isolates from effluent samples showed fastest mean exponential growth with a specific growth rate of μ = 0.25 ± 0.04 ∆OD h−1 followed by biofilm (μ = 0.20 ± 0.02 ∆OD h−1) and influent (μ = 0.15 ± 0.03 ∆OD h−1). Biofilm isolates were the fastest to reach the exponential phase after ~8 h (Figure A). Effluent and influent isolates showed comparable growth dynamics, with exponential growth starting at ~12 h after incubation. This is not surprising since both originate from the same cooling-lubricating stream, but before and after the sedimentation basin. However, isolates from the effluent were the fastest to reach the stationary phase.
Figure 4. Growth experiments with 30 isolates from four sampling sites conducted without treatment. (A) Growth curves. (B) Concentration changes of TEA and bicine during exponential growth.
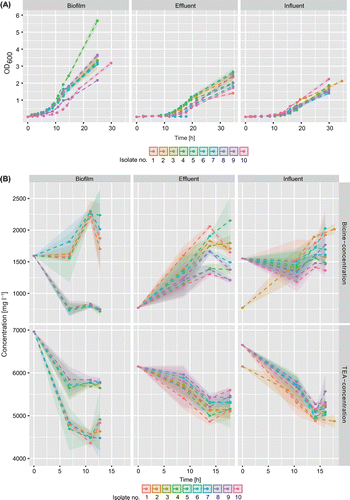
All isolates rapidly degraded TEA (Figure B). As all isolates from influent and effluent samples were identified as P. pseudoalcaligenes, the observed similarities in metabolic rates may be attributed to physiological similarity of these isolates and their close phylogenetic relationship. For isolates from biofilm samples, mean degradation of TEA was calculated for P. pseudoalcaligenes and A. faecalis. Highest degradation of TEA was observed for A. faecalis with 1,807 ± 510 mg l−1 and for P. pseudoalcaligenes with 1,616 ± 754 mg l−1 over 13 h until the stationary growth phase. In contrast to other isolates the majority of TEA was already degraded before entering the exponential growth phase at ~min400. As a consequence, degradation rates during exponential growth were lower (–3.7 ± 22.7 mg h−1 and 20.9 ± 36.5 mg h−1 for A. faecalis and P. pseudoalcaligenes, respectively). Measurements started at the beginning of the lag phase and were not measured in short intervals, which can explain the low resolution of the initial degradation progress. For isolates from effluent, degradation started more slowly and led to a mean decrease of 856 ± 175 mg l-1 during the 18 h incubation period, with degradation rates of 48.5 ± 18.2 mg h−1 during exponential growth. Isolates from influent samples exhibited the highest mean degradation rates during log phase (79.6 ± 40.7 mg h-1), with an overall degradation of 1,413 ± 161 mg l-1 over a time-span of 16 h. After an initial degradation of TEA, the slight increase in the TEA concentration found towards the end of exponential growth in some samples may be attributed to the high variance found among samples. Moreover, some isolates seem to be capable of bicine degradation. This relationship between TEA degradation and bicine production is best illustrated in effluent samples. In this case a trend change was visible in the latter stage of the exponential phase. Except for degradation rates, there were no other differences among Pseudomonas isolates. The highest mean increase in bicine in the sampled timeframe was observed in effluent isolates with 780 ± 307 mg l−1, followed by influent with 218 ± 415 mg l−1. The mean concentration change in biofilm cultures was calculated to be negative (–140 ± 688 mg l−1 for A. faecalis and -423 ± 732 mg l−1 for P. pseudoalcaligenes), since some isolates showed a strong decrease in bicine concentration in contrast to the remaining cultures.
The rate of bicine formation during log phase was comparable in the effluent (27.6 ± 27.6 mg h−1) and the biofilm (33.0 ± 49.4 mg h−1 for A. faecalis and 11.2 ± 19.8 mg h−1 for P. pseudoalcaligenes). Rates of bicine formation in influent isolates, however, were approximately twice as high, at 55.2 ± 25.8 mg h−1. When the results from the growth and degradation records were linked, isolates deriving from influent were fastest to degrade TEA and increase bicine concentration during exponential growth. Biofilm isolates, however, exhibited the highest capacity regarding overall TEA degradation.
Growth experiments with alkaline and heat treatment
Growth experiments under modified conditions were conducted in CLM to investigate easy-to-install solutions for retarded bacterial degradation of TEA in the industrial CLC. Three isolates of A. faecalis and two isolates of P. pseudoalcaligenes with fast growth were selected from the biofilm isolates and grown under pH 9 and after initial exposure to 50°C for 5 and 15 min, respectively. Exposure to 50°C for 15 min postponed the onset of exponential growth (Figure A) by 25 to 48 h. The mean specific growth rate in both the temperature treatment groups was similar with μ = 0.13 ± 0.05 ∆OD h−1 for a 5 min exposure time and μ = 0.12 ± 0.005 ∆OD h−1 for a 15 min exposure time..
Figure 5. (A) Growth curves of five selected biofilm isolates (1, 2, 5: Alcaligenes faecalis; 5, 7: Pseudomonas pseudoalcaligenes) grown at 50°C for 5 or 15 min or that were grown under elevated pH compared to the control. (B) Concentration changes of TEA and bicine during exponential growth.
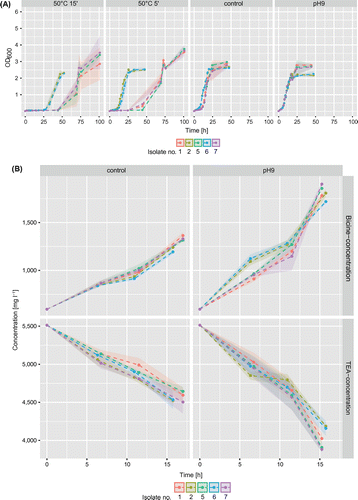
In contrast to the microtoxic pH effect described in Bakalova et al. (Citation2014), the application of a higher pH in the medium led to faster growth and higher specific growth rate (μ = 0.21 ± 0.01 ∆OD h−1) compared to the control (μ = 0.16 ± 0.04 ∆OD h−1). Probably, the elevated pH had no inhibiting potential because the selected isolates of Alcaligenes sp. and Pseudomonas sp. were tolerant to high pH and adapted to TEA (Thangam and Rajkumar Citation2000; Luque-Almagro et al. Citation2005).
Measurements of TEA and bicine concentrations were only performed for the control and the pH 9 treated group, as the thermal treatment only postponed microbial growth but did not change the exponential growth rates. At the higher pH of 9 compared to the usual pH of 8 in the industrial cooling-lubricating circuit, TEA degradation rates were elevated with a simultaneous strong increase in bicine concentration (Figure B). Mean degradation rates were calculated from all five isolates, as no large deviation was observed (variations of <5% for species-segregated means compared to the overall means). In cultures with elevated pH, TEA degradation rates during exponential growth were 105 ± 25 mg h−1, which is nearly twice as high as in the controls (54.1 ± 5.8 mg h-1). Similarly, the bicine increase under pH 9 amounted to 91.6 ± 20.9 mg h-1 while the controls showed an increase of 43.4 ± 4.0 mg h-1 during log phase. In the observed 17 h timeframe, a mean overall TEA degradation rate of 1,481 ± 140 mg l−1 was observed in the pH 9 group, while it was only 954 ± 58 mg l−1 in the control. In line with TEA degradation, the bicine increase in cultures with elevated pH was nearly twice as high (1,219 ± 70 mg l−1) than in the controls (695 ± 70 mg l−1).
This study suggests that the implementation of a heating step represents a potential way to delay growth and therefore reduce TEA degradation in industrial cooling liquid circuits. A rise in the temperature of the medium to 50°C for 15 min was sufficient to delay the transition from lag to exponential phase for up to 45 h.
Since only selected isolates from biofilm samples were used for the growth experiments this finding would need to be confirmed under more realistic conditions similar to those in the real CLC. Further improving the experimental procedure, the biofilm isolates should be grown in a biofilm-based system like a flow cell, and additional biofilm control strategies like enzymes, phages, and antimicrobial molecules from competing species (Simões et al. Citation2010) should be considered.
In conclusion, in order to control the growth of TEA degraders in cooling liquid circuit of industrial plants, the bacterial community at different sites was identified and TEA degraders were isolated and further analyzed. The genera Pseudomonas and Clostridium accounted for the highest abundance at all sampling sites. The sites contained different microbial communities, although all parts of the cooling circuit were interconnected and continuously in contact with the cooling liquid. These differences can be attributed to structural differences in the circuit, such as the degree of oxygen concentration, contact to production processes, the intensity of the cooling liquid contact as well as exposure to purification measures like flocculation tanks and sedimentation basins. The latter was shown to have a considerable impact on bacterial abundances and reduced microbial load in subsequent sections of the cooling circuit by up to 80%. Isolates capable of TEA degradation were identified as P. pseudoalcaligenes and A. faecalis. The growth experiments showed a considerable potential for the modification of process conditions to offset microbial degradation of TEA. Although the present results hint towards a significant contribution of biofilms to TEA degradation, it will be necessary to estimate microbial biomass and activity in biofilms and the suspension throughout the CLC and to assess process rates not only in the liquid culture but also in biofilms.
Conflict of interest disclosure statement
The authors declare no conflict of interests.
Funding
This project was funded by the Nachwuchsförderung of the Leopold-Franzens-Universität Innsbruck.[award id: 172768]
Supplemental data
The supplemental material for this paper is available online at https://doi.org/10.1080/08927014.2018.1468887
Supplemental Material
Download PDF (133.7 KB)References
- Adapa LM, Azimi Y, Singh S, Porcelli D, Thompson IP. 2016. Comparative study of chemical and physical methods for distinguishing between passive and metabolically active mechanisms of water contaminant removal by biofilms. Water Res. 101:574–581. doi:10.1016/j.watres.2016.06.015.
- Auguet O, Pijuan M, Borrego CM, Rodriguez-Mozaz S, Triadó-Margarit X, Giustina SV, Gutierrez O. 2017. Sewers as potential reservoirs of antibiotic resistance. Sci Total Environ. 605-606:1047–1054. doi:10.1016/j.scitotenv.2017.06.153.
- Bakalova S, Mincheva V, Doycheva A, Groudeva V, Dimkov R. 2014. Microbial toxicity of ethanolamines. Biotechnol Biotechnol Equip. 22:716–720. doi:10.1080/13102818.2008.10817540.
- Benedek T, Táncsics A, Szabó I, Farkas M, Szoboszlay S, Fábián K, Maróti G, Kriszt B. 2016. Polyphasic analysis of an Azoarcus-Leptothrix-dominated bacterial biofilm developed on stainless steel surface in a gasoline-contaminated hypoxic groundwater. Environ Sci Pollut Res. 23:9019–9035. doi:10.1007/s11356-016-6128-0.
- Bizet J, Bizet C. 1997. Strains of Alcaligenes faecalis from clinical material. J Infect. 35:167–169. doi:10.1016/S0163-4453(97)91710-2.
- Bodenhofer U, Bonatesta E, Horejs-Kainrath C, Hochreiter S. 2015. msa: an R package for multiple sequence alignment. Bioinformatics. 31:3997–3999. doi:10.1093/bioinformatics/btv494.
- Buers KLM, Prince EL, Knowles CJ. 1997. The ability of selected bacterial isolates to utilise components of synthetic metal-working fluids as sole sources of carbon and nitrogen for growth. Biotech Lett. 19:791–794. doi:10.1023/A:1018300628816.
- Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 27:2194–2200. doi:10.1093/bioinformatics/btr381.
- Eide-Haugmo I, Brakstad OG, Hoff KA, Sørheim KR, da Silva EF, Svendsen HF. 2009. Environmental impact of amines. Energy Procedia. 1:1297–1304. doi:10.1016/j.egypro.2009.01.170.
- Flemming H-C, Wingender J. 2010. The biofilm matrix. Nat Rev Microbiol. 8:623–633. doi:10.1038/nrmicro2415.
- Frings J, Wondrak C, Schink B. 1994. Fermentative degradation of triethanolamine by a homoacetogenic bacterium. Arch Microbiol. 162:103–107. eng. doi:10.1007/BF00264381.
- Fytianos G, Ucar S, Grimstvedt A, Hyldbakk A, Svendsen HF, Knuutila HK. 2016. Corrosion and degradation in MEA based post-combustion CO2 capture. Int J Greenhouse Gas Control. 46:48–56. doi:10.1016/j.ijggc.2015.12.028.
- Glasser NR, Kern SE, Newman DK. 2014. Phenazine redox cycling enhances anaerobic survival in Pseudomonas aeruginosa by facilitating generation of ATP and a proton-motive force. Mol Microbiol. 92:399–412. doi:10.1111/mmi.2014.92.issue-2.
- Grant DM, Bott TR. 2005. Biocide dosing strategies for biofilm control. Heat Transfer Eng. 26:44–50. doi:10.1080/01457630590890166.
- Jia R, Tan JL, Jin P, Blackwood DJ, Xu D, Gu T. 2018. Effects of biogenic H2S on the microbiologically influenced corrosion of C1018 carbon steel by sulfate reducing Desulfovibrio vulgaris biofilm. Corros Sci. 130:1–11. doi:10.1016/j.corsci.2017.10.023.
- Knapp JS, Jenkey ND, Townsley CC. 1996. The anaerobic biodegradation of diethanolamine by a nitrate reducing bacterium. Biodegradation. 7:183–189. doi:10.1007/BF00058178.
- Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. 2013. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol. 79:5112–5120. doi:10.1128/AEM.01043-13.
- Lawson GL, Cummings AL, Mecum S. 2003. Amine plant corrosion reduced by removal of bicine. Proceedings of the Gas Processors Association Annual Convention; Mar 12; San Antonio, TX, USA, GPA Midstream Association.
- Lessmann H, Uter W, Schnuch A, Geier J. 2009. Skin sensitizing properties of the ethanolamines mono-, di-, and triethanolamine: data analysis of a multicentre surveillance network (IVDK) and review of the literature. Contact Dermatitis. 60:243–255. doi:10.1111/j.1600-0536.2009.01506.x.
- Libralato G, Volpi Ghirardini A, Avezzù F. 2010. Seawater ecotoxicity of monoethanolamine, diethanolamine and triethanolamine. J Hazard Mater. 176:535–539. doi:10.1016/j.jhazmat.2009.11.062.
- Liu Y, Zhang W, Sileika T, Warta R, Cianciotto NP, Packman A. 2009. Role of bacterial adhesion in the microbial ecology of biofilms in cooling tower systems. Biofouling. 25:241–253. doi:10.1080/08927010802713414.
- Loy A, Lehner A, Lee N, Adamczyk J, Meier H, Ernst J, Schleifer K-H, Wagner M. 2002. Oligonucleotide microarray for 16S rRNA gene-based detection of all recognized lineages of sulfate-reducing prokaryotes in the environment. Appl Environ Microbiol. 68:5064–5081. doi:10.1128/AEM.68.10.5064-5081.2002.
- Luque-Almagro VM, Huertas M-J, Martinez-Luque M, Moreno-Vivian C, Roldan MD, Garcia-Gil LJ, Castillo F, Blasco R. 2005. Bacterial degradation of cyanide and its metal complexes under alkaline conditions. Appl Environ Microbiol. 71:940–947. doi:10.1128/AEM.71.2.940-947.2005.
- Macherey-Nagel. 2014/Rev. 04. Genomic DNA from soil - user manual: NucleoSpin(r) soil. Düren, Germany, Macherey-Nagel GmbH.
- Mattsby-Baltzer I, Sandin M, Ahlstrom B, Allenmark S, Edebo M, Falsen E, Pedersen K, Rodin N, Thompson RA, Edebo L. 1989. Microbial growth and accumulation in industrial metal-working fluids. Appl Environ Microbiol. 55:2681–2689.
- Nagler M, Podmirseg SM, Griffith GW, Insam H, Ascher-Jenull J. 2018. The use of extracellular DNA as a proxy for specific microbial activity. Appl Microbiol Biotechnol. 102:2885–2898. doI:10.1007/s00253-018-8786-y.
- Oksanen J, Guillaume Blanchet F, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O’Hara RB, Simpson GL, Solymos P, et al. 2016. vegan: community ecology package.: R package version 2.4-1. [Internet]. Available from: https://cran.r-project.org/package=vegan.
- Prince EL, McCabe RW, Morton LHG. 1994. The use of 1H nuclear magnetic resonance spectroscopy and gas liquid chromatography/mass spectrometry to determine the effects of fungal biodeterioration on defined synthetic metal-working fluids. Int Biodeterior Biodegradation. 34:1–20. doi:10.1016/0964-8305(94)90017-5.
- R Development Core Team. 2008. R: a language and environment for statistical computing [Internet]. Vienna, Austria: [publisher unknown]. ISBN: 3-900051-07-0; [cited 2017 Feb 16]. Available from: http://www.r-project.org/.
- Roberts DW. 2016. labdsv: ordination and multivariate analysis for ecology: R package version 1.8-0. [Internet]. Available from: https://cran.r-project.org/package=labdsv.
- Sandin M, Allenmark S, Edebo L. 1990. Selective toxicity of alkanolamines. Antimicrob Agents Chemother. 34:491–493. doi:10.1128/AAC.34.3.491.
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, et al. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 75:7537–7541. doi:10.1128/AEM.01541-09.
- Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. 2011. Metagenomic biomarker discovery and explanation. Genome Biol. 12:R60.10.1186/gb-2011-12-6-r60.
- Simões M, Simões LC, Vieira MJ. 2010. A review of current and emergent biofilm control strategies. LWT-Food Sci Technol. 43:573–583. doi:10.1016/j.lwt.2009.12.008.
- Simões LC, Lemos M, Pereira AM, Abreu AC, Saavedra MJ, Simões M. 2011. Persister cells in a biofilm treated with a biocide. Biofouling. 27:403–411. doi:10.1080/08927014.2011.579599.
- Sinai L, Rosenberg A, Smith Y, Segev E, Ben-Yehuda S. 2015. The molecular timeline of a reviving bacterial spore. Mol Cell. 57:695–707. doi:10.1016/j.molcel.2014.12.019.
- Speranza G, Morelli CF, Cairoli P, Müller B, Schink B. 2006. Mechanism of anaerobic degradation of triethanolamine by a homoacetogenic bacterium. Biochem Biophys Res Commun. 349:480–484. doi:10.1016/j.bbrc.2006.08.001.
- Teixeira P, Oliveira R. 2002. Metabolism of Alcaligenes denitrificans in biofilm vs planktonic cells. J Appl Microbiol. 92:256–260. doi:10.1046/j.1365-2672.2002.01525.x.
- Thangam EB, Rajkumar GS. 2000. Studies on the production of extracellular protease by Alcaligenes faecalis. World J Microbiol Biotechnol. 16:663–666. doi:10.1023/A:1008989810481.
- Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680. eng. doi:10.1093/nar/22.22.4673.
- Toyofuku M, Inaba T, Kiyokawa T, Obana N, Yawata Y, Nomura N. 2015. Environmental factors that shape biofilm formation. Biosci Biotechnol Biochem. 80:7–12. doi:10.1080/09168451.2015.1058701.
- Weavers LK, Hua I, Hoffmann MR. 1997. Degradation of triethanolamine and chemical oxygen demand reduction in wastewater by photoactivated periodate. Water Environ Res. 69:1112–1119. doi:10.2175/106143097X125849.
- Wickham H. 2009. ggplot2. New York (NY): Springer; p. 212. doi:10.1007/978-0-387-98141-3.
- Yilmaz P, Parfrey LW, Yarza P, Gerken J, Pruesse E, Quast C, Schweer T, Peplies J, Ludwig W, Glöckner FO. 2014. The SILVA and “All-species Living Tree Project (LTP)” taxonomic frameworks. Nucleic Acids Res. 42:D643–D648. doi:10.1093/nar/gkt1209.