ABSTRACT
The objective of this study was to investigate the genetic polymorphisms and transcript levels of antioxidant and metabolic markers for diarrhea monitoring in Holstein dairy calves. For DNA and RNA extraction, blood samples were taken from 100 female calves (50 with diarrhea and 50 appearing healthy). DNA sequencing and real time PCR for and metabolic genes were performed to identify genetic variants linked to diarrhea susceptibility. Results revealed that there were nucleotide sequence variations in the investigated genes between healthy and diarrheal calves. Keap1, HMOX1, CMPK2, ASPG, FPGT, TNNI3 K, and LPCAT1 were considerably more highly expressed in diarrheic calves than resistant ones. The Nrf2, PRDX2, and PRDX6 genes, however, produced a different pattern. This study provides a practical control strategy to lessen newborn diarrhea in Holstein dairy calves and emphasizes the significance of nucleotide variations along with antioxidant and metabolic gene expression patterns as proxy markers of diarrhea susceptibility/resistance.
Introduction
Calves are essential to preserving animal wealth because they act as a herd replacement or as a necessary source of high-quality protein to satisfy the demands of a population that is quickly growing (Zaki, Citation2003). Improved understanding of the biology of the digestive tract in newborn dairy calves will lead to a reduction in health problems such diarrhoea as well as an increase in feed efficacy and average daily growth in pre-weaned dairy calves (Rosa & Osorio, Citation2019). There were moderate to high incidences of diarrhea, particularly in the Holstein breed (Becher et al., Citation2004). Calves up to 30 days old are the primary age group affected by enteric disorders (Cho & Yoon, Citation2014).
Calf diarrhoea (CD), one of the most common problems with young animals, has a huge detrimental economic and production impact on the global cattle industry (Achá et al., Citation2004). Although farm management has been optimized, including calf feeding techniques alongside advancements in calf husbandry systems, neonatal calf diarrhea (NCD) still accounts for between 27.4% and 55% of all young calf deaths in Egypt (Younis et al., Citation2009). In addition to mortality, economic losses also occur from remedy, diagnostics, labor interpolation, and reduced herd size, as well as succeeding prolonged illness and poor growing performance, also result in economic losses (Bazeley, Citation2003).
NCD is a complex syndrome that also involves non-infectious problems with the cattle (such as food and immune status), the management, or the environment (Izzo et al., Citation2011). Effective control of NCD is challenging due to its multifactorial character (Cho & Yoon, Citation2014). Diarrhea instances are typically more common from October/November to March, when it rains, than from March to October (Achá et al., Citation2004). Despite the complicated causes of calf diarrhea, bacterial organisms are still to blame for more than 50% of cases in newborn calves (Kumar et al., Citation2012). Antibiotic use may lead to the evolution of antibiotic resistance, risking both the general public's health and the ability to treat bacteria that cause calf diarrhea in the future. Alternatives to antibiotics are desperately required for disease control and prevention. It has been suggested that it may be possible to stop the virulence factors that the bacteria use to produce diarrhea (Rasko and Sperandio, Citation2010).
The intestine is the main organ of the immune system. Given that more than 70% of immune system cells are found in the gastrointestinal tract; where there is a higher concentration of immune cells than in any other body tissue, making it a strong predictor of an animal's health when it is young (Wu & Wu, Citation2012). Redox has become a key mode of chemical signaling in the intestine, and it is particularly susceptible to oxidative stress in immune cells (Cheng et al., Citation2021). Reactive oxygen (ROS) concentrations inside of cells that are higher than physiologic levels cause oxidative stress, which in turn triggers oxidative injury and an inflammatory response in the gut that eventually results in diarrhea (Bhattacharyya et al., Citation2014). Newborns are particularly susceptible to perinatal diseases, which have high mortality rates, because of their metabolic instability (Ramel et al., Citation2014). Breeders can find genetic indicators for diarrhea susceptibility by identifying antioxidant and metabolic potential genes, as well as their mutations that alter gene expression and phenotype (Hassan et al., Citation2002).
Marker aided selection (MAS) is a method for finding genetic factors that influence susceptibility to common diseases (Bishop et al., Citation1995; Ashwell et al., Citation1997). In the time of genomic selection, the implementation of selection approaches for novel functional qualities, such as disease resistance, is made possible by great cow training sets merging phenotypes with high-throughput genomic SNP indicator information (Buch et al., Citation2012). The significant shortening of generational intervals is a second significant benefit of genetic selection (Schaeffer, Citation2006). According to this viewpoint, characteristics identified early in life, such as calf traits, may serve as useful early indicators of future health or cow output. Diarrhea in calves has an impact on the animal's performance and productivity in later life. The likelihood of heifer health disorders is typically increased when a calf illness occurs (Sivula et al., Citation1996). According to Warnick et al. (Citation1994) dystocia at first calving was phenotypically associated with calf respiratory and diarrhea. The term ‘transcriptome’ denotes all the genes in the genome that are accurately and at high throughput during particular physiological and pathological states (Kukurba & Montgomery, Citation2015). It is commonly applied to immune monitoring in inflammatory disorders to uncover pathogenic, diagnostic, and prognostic indicators, and it is essential in the detection of novel diagnostic or therapeutic goals (Tang et al., Citation2011; Banchereau et al., Citation2017).
Finding genetic pathways with varied manifestations of disease resistance is now being done in livestock genomics using the candidate gene technique (Feuk et al., Citation2006). Utilizing genomic technology has enhanced molecular genetics and opened up intriguing possibilities for the identification of useful genes. Thousands of single nucleotide polymorphisms (SNPs) have been found in the genomes of various livestock animals thanks to genome sequencing programs (Tsuchida et al., Citation2010). These genetic markers, or SNPs, can be used to identify the genetic variation that underlies traits in livestock animals that are economically significant and to better understand how genetic variants connect to various phenotypes (Ateya et al., Citation2023). Our understanding of the genetic pathways and genetic basis of numerous phenotypic traits in different species, including cattle, has significantly improved in recent years as a result of the development of affordable sequencing technology and huge data processing tools (Essa et al., Citation2023). In order to make it easier to choose breeding animals that will actually boost disease resistance traits, genetically based improvement programs should be created.
As there is little information on the antioxidant and metabolic pathways that determine a Holstein dairy calf's resistance/susceptibility to diarrhea, it is important to note that more research is necessary to follow prospective indicators that might suggest mortality and indisposition in neonatal calves from diarrhea (Perez et al., Citation1990; Gulliksen et al., Citation2009). The molecular alterations may also help to diagnose diarrhea and provide decisive information about the interactions between the physiology of the intestines, antioxidants, and metabolic pathways (Klein-Jöbstl et al., Citation2015). This study used PCR-DNA sequencing and real-time PCR approaches to examine potential antioxidant and metabolic genes efficacy as candidates for prediction and tracking diarrhea resistance/susceptibility in Holstein dairy calves.
Materials and methods
Dairy calves and experimental samples
This research used 100 female Holstein dairy calves upraised on a private farm in the province of Ismailia, Egypt, from November 2022 to January 2023, of which 50 were diarrheal and 50 appeared to be in good health. The calves were 45 days old, weighed 69 ± 4.5 kg, and were fed whole milk. After giving birth, calves were separated from their mothers and moved to different facilities where they received colostrum through feeding tubes. A colostrometer was used to gauge the grade of the colostrum, and colostrum (1035 mg/ml) with a high specific weight was provided to the calves. Within the first 24 h, calves received about 5 liters of colostrum, after which they received milk twice daily for the following days. All calves were examined by the same veterinarian each day. The diarrheic calves were selected on the basis of physical examination during the early stages of the condition, giving close attention to body temperature. The studied claves experienced a medical inspection that involved instantaneous recording of their body's temperature, pulse, respiration rate, mucous membranes, and faeces’ consistency and color (Radostits et al., Citation2010). The jugular vein of every calf were punctured to attain five milliliters of blood. In order to recover DNA and RNA, the samples were positioned into tubes filled with anticoagulants in a vacuum to acquire whole blood (EDTA or sodium fluoride). All animal management procedures, tested trials collection, and sample discarding were carried out underneath the supervision of the University of Sadat City's Veterinary Medical school in accordance with IACUC guidelines (Code VUSC-006-1-23).
Isolation and amplification of DNA
By means of the genetic material JET entire blood genomic DNA isolation kit and the manufacturer's guidelines, total blood was used to extract the genome's DNA (Thermo scientific, Vilnius, Lithuania). Using Nanodrop, DNA of high purity and concentration was analyzed. The following genes for antioxidants (Nrf2, Keap1, PRDX2, PRDX6, and HMOX1) and metabolic (CMPK2, ASPG, FPGT, TNNI3K and LPCAT1) were amplified. The Bos taurus genome that was accessible in PubMed was employed to create the oligonucleotide sequences for amplification. contains a list of the primers used during the PCR.
Table 1. Oligonucleotide primer of Antioxidant and metabolic genes employed for genetic polymorphisms.
A heat cycler has a 150 mL final volume was used to process the polymerase chain amplification mixture. Each reaction container contained the following components: 66 μL d.d. water, DNA with 6 microliters, each matching primer with 1.5 microliters, and of master mixture with 75 microliters (Jena Bioscience, Jena, Germany). The PCR combinations spent four minutes at a starting temperature of 95 °C for unwinding. The 35-cycles included 95 °C denaturation cycles lasting one minute each, annealing cycles lasting one minute at the temps listed in , 30-second rounds for elongation at 72 °C, ten additional minutes of extending occurred at 72 °C. The materials were saved at 4 °C. A gel certification system was employed to find demonstrative PCR findings using agarose gel electrophoresis and to view PCR segment patterns under UV light.
Finding polymorphism in antioxidant and metabolic genes
Prior to DNA sequencing, Jena Bioscience # pp-201s/Munich, Hamburg, Germany, offered tools for purifying PCR and eliminate primer dimmers, non-specific bands, and other contaminants, producing the intended amplified product of the predicted scope (Boom et al., Citation1990).
Using a Nanodrop (Waltham, Massachusetts, USA, UV-Vis spectrometer Q5000), satisfactory quality and good concentrations were obtained for measuring PCR output (Boesenberg-Smith et al., Citation2012). Healthy alongside diarrheal heifers have been used to search for SNPs using sequencing of the amplified PCR products. The PCR yields were sequenced on an ABI 3730XL DNA sequencer (United States: Applied Biosystems, Waltham) operating the Sanger et al., Citation1977 described enzyme chain terminator method.
The software programs Chromas 1.45 and BLAST 2.0 accustomed for examining the DNA sequence analysis outcomes (Altschul et al., Citation1990). Polymorphisms have been identified when comparing the antioxidant and metabolic genes product produced by PCR to the reference gene sequences obtained from GenBank. Relying on the sequence matching amongst the dairy calves, the MEGA4 tool has the ability to recognize dissimilarities in the examined genes’ amino acid sequences (Tamura et al., Citation2007).
Transcript levels of antioxidant and metabolic genes
Following the manufacturer's instructions, the whole RNA was extracted from the blood samples taken from the investigated dairy heifers using the Trizol solution (RNeasy Mini Ki, 74104, Product No.). We measured and verified the quantity of the isolated RNA using a NanoDrop® ND-1000 spectrophotometer. The complementary nucleic acid for each sample has been produced using the producer's method (Waltham, Massachusetts, USA: Thermo Fisher, Product No. EP0441). SYBR Green PCR Master Mix and quantifiable RT–PCR have been employed to evaluate the expression profiles of the antioxidants (Nrf2, Keap1, PRDX2, PRDX6, and HMOX1) and metabolic (CMPK2, ASPG, FPGT, TNNI3K and LPCAT1) genes (2x SensiFastTM SYBR, Bio-line, CAT No: Bio-98002). The SYBR Green PCR Master Mix have been exploited for computing comparative amount possessed by the mRNA (Toronto, Ontario, Canada: Quantitect SYBR green PCR reagent, Catalog No. 204141).
Using the Bos taurus genome from PubMed, the sense and antisense primer sequences were generated (). The ß. actin gene served as the constitutive reference. Overall RNA with 25 microliters, 1 microliter of every matching primer, 8 microliters of water without Nuclease, 0.5 microliter of inverse transcriptase, 12.5 microliters of Quantitect SYBR green reaction master solution, and 3 microliters of Trans Amp buffer made up the PCR combination. The finished reaction mixture then underwent the following steps inside a thermal cycler: inverse transcription for 30 min at 55 °C; preliminary denaturation aimed at 8 min at 95 °C; 40 cycles at 95 °C aimed at 15 s and the primer binding temperatures specified throughout ; and extending aimed at 1 min at 72 °C. A melting curve investigation was employed subsequent to the amplifying step for proving specificity of the amplified product. By comparing each gene's expression in the analyzed sample to that of the ß. Actin gene, the 2−ΔΔCt scheme has been exploited for considering the differences in the expression of each gene (Livak & Schmittgen, Citation2001; Pfaffl, Citation2001).
Table 2. Oligonucleotide forward and reverse primers for antioxidant and metabolic genes under investigation used in real time PCR.
Statistical analysis
Ho: Exploring genetic polymorphisms and transcript levels of antioxidant and metabolic markers could not predict and monitor diarrhea in Holstein dairy calves.
HA: Exploring genetic polymorphisms and transcript levels of antioxidant and metabolic markers could predict and monitor diarrhea in Holstein dairy calves.
The substantial differences in the discovered genes’ SNPs between the examined calves were found using a chi-square analysis. A statistical investigation have been exploited for this reason using the Graphpad statistical program (Graphpad prism for Windows version 5.1, Graphpad Software, Inc., San Diego, CA, USA) (p < 0.05). The Statistical Package for Social Science (SPSS) version 17 computer program and the t-test have been exploited for judging if it was present a statistically noteworthy variance between healthy and diarrheal calves. Mean and standard error (Mean ± SE) have been used to present the findings. The significance of the variations was assessed using p < 0.05.
Results
Genetic polymorphisms of antioxidant and metabolic genes
Healthy and affected dairy calves in good health and those who had diarrheal disease had different SNPs in the amplified DNA bases, according to the results of the PCR-DNA sequencing for the Nrf2 (340-bp), Keap1 (515-bp), PRDX2 (315-bp), PRDX6 (457-bp), HMOX1 (489-bp), CMPK2 (536-bp), ASPG (339-bp), FPGT (471-bp), TNNI3K (358-bp) and LPCAT1 (468-bp) genes. All the discovered SNPs were approved using the DNA sequence differences between antioxidant and metabolic markers investigated in the researched heifers and the reference gene sequences obtained from GenBank (Supplemental Figures S1–S10). shows the dissemination of a single base variation as well as a type of inherited change for antioxidant and metabolic indicators in non-diarrheic and diarrheal dairy calves. According to the chi-square analysis of the SNPs, the healthy and diarrheal calves did demonstrate noticeably different occurrences of the investigated markers (p < 0.05). The coding DNA sequences of the diarrheal calves differed from those of healthy calves due to the exonic region mutations. Eleven SNPs were discovered using DNA sequencing of antioxidant genes; 10 of them are non-synonymous and one is synonymous. The DNA sequencing of metabolic genes revealed eight SNPs, 5 of which are non-synonymous and 3 of which are synonymous.
Table 3. Single base difference dispersal as well as sort of inherited change for antioxidant and metabolic markers in non-diarrheic and diarrheic dairy calves.
Patterns for transcript levels of antioxidant and metabolic indicators
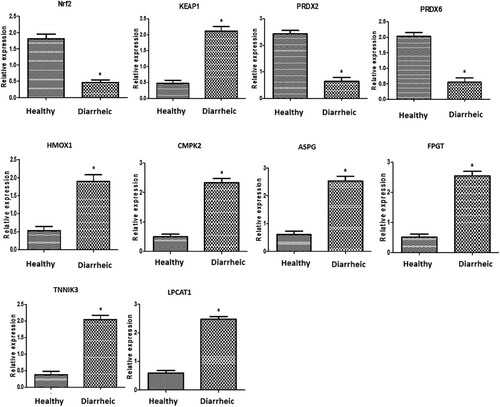
displays the measured antioxidant and metabolic transcript patterns. The Nrf2, PRDX2, and PRDX6 genes were expressed at substantially lower levels in diarrheal calves (P < 0.05). However, Keap1, HMOX1, CMPK2, ASPG, FPGT, TNNI3 K, and LPCAT1 evoked an opposite trend. For each gene examined in the diarrheic heifers, FPGT had the highest potential mRNA level (2.54 ± 0.15); Nrf2 had the lowest potential level (0.46 ± 0.07). In the healthy calves, PRDX2 had the highest potential amount of mRNA (2.43 ± 0.12), while TNNIK3 had the lowest (0.37 ± 0.11).
Discussion
Diarrhea is one of the normal symptoms of disease in animals. Strong emission or reduced absorption from the gastrointestinal layer are physiological processes caused by a variety of factors (Semrad, Citation2012). When GIT peristalsis increases its reaction to an upsurge for intestine fluid, thin faeces are excreted. Sometimes diarrhea is a digestive defense mechanism, but more often it is caused by bacteria that inflame the digestive mucosa and cause it to release excessive amounts of pus, blood, and infrequently mucus (Lundgren & Svensson, Citation2001; Kaper et al., Citation2004). Although there is a wealth of scientific information regarding newborn ruminant livestock morbidity and mortality, this knowledge did not significantly improve survival (von Buenau et al., Citation2005; Celi et al., Citation2017). The reason may be because studies have not focused much on newborn morbidity, preferring to uncover and assess remedies to issues brought on by economic concerns (von Buenau et al., Citation2005). For enhancing animal wellbeing, finding more powerful potential biomarkers is important for the diagnosis of neonatal diseases (Nataro and Kaper, Citation1998).
An understanding of the genes, the underlying mutations, and the interactions with other factors that impart resistance is required for the efficient exploitation of disease-resistant livestock or the total eradication of diseased livestock (Pal & Chakravarty, Citation2020). The antioxidants (Nrf2, Keap1, PRDX2, PRDX6, and HMOX1) and metabolic (CMPK2, ASPG, FPGT, TNNI3K and LPCAT1) genes in diarrhea-affected and healthy Holstein dairy calves were characterized in this research using a PCR-DNA sequencing technique. The findings show that the SNPs involving both categories vary. The chi-square study revealed that nucleotide polymorphism dispersion amongst the inspected calves was significant (p < 0.05). It is important to emphasize that the polymorphisms found and made available in this context give additional data for the evaluated indicators when compared to the corresponding datasets acquired from GenBank.
There have been recent studies targeting novel genes specific to livestock diarrhea susceptibility using genome-wide association analysis (Casas et al., Citation2015; Kirkpatrick et al., Citation2022), but up to this point no studies have examined the link between the SNPs in these genes and diarrhea risk. We are the first to show this connection using the PubMed European cow (Bos taurus) gene sequences. According to our knowledge, there hasn't been any prior research on the variation of the antioxidants (Nrf2, Keap1, PRDX2, PRDX6, and HMOX1) and metabolic (CMPK2, ASPG, FPGT, TNNI3K and LPCAT1) markers and how they relate to diarrhea in Holstein dairy calves. The candidate gene method, however, has been employed to keep track of the soundness of diarrheal newly born animals. Dairy heifers, for example, did not demonstrate any association between clinical intestinal disorders and CXCR1 SNPs (Tsuchida et al., Citation2010). TLR4 gene polymorphisms was associated with diarrhea onset in dairy calves (Judi et al., Citation2020). The DRA gene for leukocyte antigen has also been related to genetic variation in piglet diarrhea (Yang et al., Citation2014). The relatedness between polymorphism of the Nramp1 gene and swine diarrhea has also been studied (Chen et al., Citation2021).
The term ‘transcriptome’ states that the genome's complete set of genes that are reliably and efficiently expressed in various physiological and pathological conditions (Kukurba & Montgomery, Citation2015). It is commonly used to assess the immune system in inflammatory disorders to locate pathogenic, diagnostic, and prognostic signs, and it has aided in the discovery of new therapeutic or diagnostic targets (Burel et al., Citation2019). Through measuring the mRNA levels of antioxidants (Nrf2, Keap1, PRDX2, PRDX6, and HMOX1) and metabolic (CMPK2, ASPG, FPGT, TNNI3K and LPCAT1) genes, we examined the changes in the redox and metabolic state in diarrheal dairy calves compared with healthy ones. The Nrf2, PRDX2, and PRDX6 genes were expressed at significantly lower levels in diarrheal calves, according to the molecular changes. However, the expression levels of the genes Keap1, HMOX1, CMPK2, ASPG, FPGT, TNNI3K, and LPCAT1 were significantly greater in diarrheal calves than in resistant ones. Gene expression as well as genomic SNP markers were employed to evaluate genetic polymorphisms in order to address the limitations of earlier studies. The mechanisms that were investigated to regulate the antioxidant and metabolic indicators in both healthy and diarrheic calves are thus widely acknowledged. This study is the first to comprehensively analyze the transcript levels of the antioxidant and metabolic indicators connected to the risk of calf diarrhea. When related to the pattern of marker expression in goats, bovine viral diarrhea virus-2-infected peripheral blood mononuclear cells mRNA investigation showed discrete activation of immunity genes (Li et al., Citation2019). Furthermore, the downstream signaling networks of TLR4 are similar in healthy and diarrheal neonatal goats (Cheng et al., Citation2021).
Antioxidants provide protection by removing or detoxifying ROS, preventing their production, or securing transition metals, which are the source of free radicals (Masella et al., Citation2005). Such mechanisms comprise both enzymatic and non-enzymatic antioxidant resistances formed within the body, known as endogenous antioxidant indicators (Glasauer & Chandel, Citation2014). The Keap1-Nrf2 stress response pathway, which regulates cytoprotective gene expression, is the primary inducible defense against oxidative stress (Baird & Yamamoto, Citation2020). Under normal conditions, Keap1 represents a substrate adaptor for cullin-based E3 ubiquitin ligase, which hinders Nrf2 transcriptional action via ubiquitination and proteasomal degradation (Kobayashi et al., Citation2004). This might explain the opposing expression pattern Keap1 and Nrf2 genes displayed in our investigation. Peroxiredoxin (PRDX) is an antioxidant enzyme oxido-reductase protein that possesses a preserved ionized thiol which allows hydrogen peroxide degradation (H2O2) (Wadleya et al., Citation2016). Heme oxygenase (HMOX), which converts heme into equimolar amounts of carbon monoxide (CO), free iron, and biliverdin, is the rate-limiting enzyme in the heme catabolic pathway (Consoli et al., Citation2021). With antioxidant, anti-inflammatory, anti-apoptotic, anti-coagulation, anti-proliferative, and vasodilator properties, it is also acknowledged as a stress-responsive protein and is postulated to perform a variety of protecting roles counter to various stresses (Mohamed et al., Citation2022).
Several regulatory enzymes of the intermediary metabolism have variable gene expression, which can offer helpful methods for enhancing genetic selection for cattle adaptation to adverse environments (van Harten et al., Citation2013). Cytidine/uridine monophosphate kinase 2 (CMPK2) was shown to be the candidate gene for immunomodulatory signaling pathways in many species (Zhang et al., Citation2020). This holds true for bacterial diseases in particular (Feng et al., Citation2021). In addition to its role in nucleotide synthesis, mitochondrial CMPK2 also takes part in DNA repair processes and the production of amino acids, which helps activated proinflammatory macrophages survive (Kanehisa et al., Citation2018). L-asparaginases, like ASPG, are essential for the biosynthesis of amino acids because they can catalyze the conversion of asparagine into aspartic acid and ammonia (Karamitros and Konrad, Citation2014). According to Butty et al., Citation2021, the ASPG gene was connected to Holstein dairy cattle's susceptibility to digital dermatitis (DD). Fucose-1-phosphate guanylyl transferase (FPGT), an essential sugar in complex carbohydrates associated with immunological responses, inflammation, and cell-to-cell communication, is a component of the L-fucose pathway (Becker & Lowe, Citation2003). The reasons of inflammation are also related to the serine/threonine protein kinase (TNNI3K) (Wiltshire et al., Citation2011). Durán Aguilar et al., Citation2017 state that the genomic region on BTA20 has been earlier recognized in Holstein and connected to SCS, which raises the possibility that its gene, Lysophospha-tidylcholine acyltransferase 1 (LPCAT1), may have an effect on the resistance to mastitis and metabolic disorders.
Because it is the primary place for the existence of several pathogens, nutrition, and communications between immune cells, the intestine is more susceptible to peroxidation than healthy tissues, which may explain the considerable change in the expression pattern of antioxidants (Nrf2, Keap1, PRDX2, PRDX6, and HMOX1) and metabolic (CMPK2, ASPG, FPGT, TNNI3K and LPCAT1) markers in diarrheal calves (Gutteridge, Citation1993; Fabiana et al., Citation2015; Wei et al., Citation2016; Aslan et al., Citation2017). Neutrophils and macrophage action may be triggered by the attack of GIT pathogens, which could be a powerful oxidizing inducer, in order to mount an immune defense against the pathogen attack. Due to the formation and accumulation of extremely large amounts of ROS, oxidative stress results (Pisani et al., Citation2009). Since the downstream signaling networks of TLR4 greatly modify method of organizing abdominal barrier purpose and could surge gut permeability to pathogens, they are obviously essential for triggering the discharge of inflammatory cytokines when there is infectious diarrhea (Fischer et al., Citation2016; Morrison et al., Citation2017; Shi et al., Citation2019). According to recent research, the amount of ROS in the colon increased along with the number of E. coli (Nathan & Cunningham-Bussel, Citation2013; Spees et al., Citation2013), which suggested that ROS additionally elaborate in the inter-bacterial antagonism. Thus, we assume that an infectious etiology is to blame for the neonatal diarrhea in the study's heifers. Our Real-Time PCR findings demonstrated a significant antioxidant response in the diarrheal calves. The ‘free radical induction hypothesis’ asserts that oxidative stress is what triggers the inflammatory process, therefore it's likely that excessive ROS formation in newborn diarrhea happened before the immunological response (Fischer et al., Citation2016). Alterations in antioxidant and metabolic indicators were also connected to the processes of tissue damage repair. Gene expression disruption can be used to characterize the common pathological processes, whereas normal gene expression controls the bulk of physiological mechanisms (Nathan & Cunningham-Bussel, Citation2013; Shi et al., Citation2019). So it should be able to investigate gene transcript levels and associated molecular pathways in order to identify and categorize the genes that produce phenotypes.
Conclusions
Single nucleotide variants (SNPs) in the genes were discovered using PCR-DNA sequencing for antioxidants (Nrf2, Keap1, PRDX2, PRDX6, and HMOX1) and metabolic (CMPK2, ASPG, FPGT, TNNI3K and LPCAT1) were found in diarrhea-free and diarrhea-infected Holstein dairy heifers. Furthermore, the levels of these indicators’ mRNA varied between healthy and diarrheal calves. These distinct functional variations provide a promising opportunity to reduce newborn diarrhea by using genetic markers along with natural resistance during cattle selection. The varying gene expression patterns for diarrhea susceptibility and resistance in calves may serve as both a guide and an indicator for assessing their health. The gene domains identified here may facilitate future approaches to the treatment of diarrhea.
Author contributions
The experimentation was developed, the PCR was completed, and the paper was written by Ahmed Ateya. Mohamed Abdo, Salah H. Faraj, Liana Fericean, and Ioan Banatean-Dunea contributed to the drafting of the manuscript. Mona Al-Sharif performed DNA sequencing and contributed to the writing of the paper. The final manuscript has been approved for publication by all writers after reading it.
Supplemental Material
Download MS Word (1.3 MB)Acknowledgments
The authors thank all members at Department of of Animal Histology and Anatomy, School of Veterinary Medicine, Badr University in Cairo (BUC), Egypt.
Data availability statement
Upon reasonable request, the supporting information for the study's findings will be provided by the corresponding author.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Achá, S. J., Kuhn, I., Jonsson, P., Mbazima, G., Katouli, M. & Mollby, R. S. (2004). Studies on calf diarrhoea in Mozambique: Prevalence of bacterial pathogens. Acta Veterinaria Scandinavica, 45, 27–36. doi: 10.1186/1751-0147-45-27
- Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. (1990). Basic local alignment search tool. Journal of Molecular Biology, 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
- Ashwell, M. S., Rexroad, Jr. C. E., Miller, R. H., Van Raden, P. M. & Da, Y. (1997). Detection of loci affecting milk production and health traits in an elite US Holstein population using microsatellite markers. Animal Genetics, 28, 216–222. doi: 10.1111/j.1365-2052.1997.00115.x
- Aslan, O., Goksu, A. G. & Apaydin, N. (2017). The evaluation of oxidative stress in lambs with Pestivirus infection. Journal of the Hellenic Veterinary Medical Society, 68, 299–306. doi: 10.12681/jhvms.15473
- Ateya, A., Al-Sharif, M., Abdo, M., Fericean, L. & Essa, B. (2023). Individual genomic loci and mRNA levels of immune biomarkers associated with pneumonia susceptibility in baladi goats. Veterinary Science, 10(3), 185. doi: 10.3390/vetsci10030185
- Baird, L. & Yamamoto, M. (2020). The molecular mechanisms regulating the KEAP1-NRF2 pathway. Molecular and Cellular Biology, 40(13), e00099-20. doi: 10.1128/MCB.00099-20
- Banchereau, R., Cepika, A. M., Banchereau, J. & Pascual, V. (2017). Understanding human autoimmunity and autoinflammation through transcriptomics. Annual Review of Immunology, 35(26), 337–370. doi: 10.1146/annurev-immunol-051116-052225
- Bazeley, K. (2003). Investigation of diarrhoea in the neonatal calf. In Practice, 25, 152–159. doi: 10.1136/inpract.25.3.152
- Becher, K. A., Robertson, I. D., Fraser, D. M., Palmer, D. G. & Thompson, R. C. A. (2004). Molecular epidemiology of Giardia and cryptosporidium infections in dairy calves originating from three sources in Western Australia. Veterinary Parasitology, 123, 1–9. doi: 10.1016/j.vetpar.2004.05.020
- Becker, D. J. & Lowe, J. B. (2003). Fucose: biosynthesis and biological function in mammals. Glycobiology, 13(7), 41–53. doi: 10.1093/glycob/cwg054
- Bhattacharyya, A., Chattopadhyay, R., Mitra, S. & Crowe, S. E. (2014). Oxidative stress: An essential factor in the pathogenesis of gastrointestinal mucosal diseases. Physiological Reviews, 94(2), 329–354. doi: 10.1152/physrev.00040.2012
- Bishop, M. D., Hawkins, G. A. & Keeler, C. L. (1995). Use of DNA markers in animal selection. Theriogenology, 43, 61–70. doi: 10.1016/0093-691X(94)00018-P
- Boesenberg-Smith, K. A., Pessarakli, M. M. & Wolk, D. M. (2012). Assessment of DNA Yield and Purity: An overlooked detail of PCR troubleshooting. Clinical Microbiology Newsletter, 34, 1–6. doi: 10.1016/j.clinmicnews.2011.12.002
- Boom, R., Sol, C. J., Salimans, M. M., Jansen, C. L., Wertheim-van Dillen, P. M. & Noordaa, J. V. D. (1990). Rapid and simple method for purification of nucleic acids. Journal of Clinical Microbiology, 28, 495–503. doi: 10.1128/jcm.28.3.495-503.1990
- Buch, L. H., Kargo, M., Berg, P., Lassen, J. & Sørensen, A. C. (2012). The value of cows in reference populations for genomic selection of new functional traits. Animal, 6, 880–886. doi: 10.1017/S1751731111002205
- Burel, J. G., Babor, M., Pomaznoy, M., Lindestam Arlehamn, C. S., Khan, N., Sette, A. & Peters, B. (2019). Host transcriptomics as a tool to identify diagnostic and mechanistic immune signatures of tuberculosis. Frontiers in Immunology, 10(19), 221. doi: 10.3389/fimmu.2019.00221
- Butty, A. M., Chud, T. C. S., Cardoso, D. F., Lopes, L. S. F., Miglior, F., Schenkel, F. S., Cánovas, A., Häfliger, I. M., Drögemüller, C., Stothard, P., Malchiodi, F. & Baes, C. F. (2021). Genome-wide association study between copy number variants and hoof health traits in Holstein dairy cattle. Journal of Dairy Science, 104(7), 8050–8061. doi: 10.3168/jds.2020-19879
- Casas, E., Hessman, B. E., Keele, J. W. & Ridpath, J. F. (2015). A genome-wide association study for the incidence of persistent bovine viral diarrhea virus infection in cattle. Animal Genetics, 46, 8–15. doi: 10.1111/age.12239
- Celi, P., Cowieson, A. J., Fru-Nji, F., Steinert, R. E., Kluenter, A. M. & Verlhac, V. (2017). Gastrointestinal functionality in animal nutrition and health: New opportunities for sustainable animal production. Animal Feed Science and Technology, 234, 88–100. doi: 10.1016/j.anifeedsci.2017.09.012
- Chen, L., Peng, S., Fu, B., Du, Q., Zhang, L. & Liu, L. (2021). Polymorphism of Nramp1 gene and its association with Diarrhea in pigs. Indian Journal of Animal Research, 55, 786–790.
- Cheng, Y., Yang, C., Tan, Z. & He, Z. (2021). Changes of intestinal oxidative stress, inflammation, and gene expression in neonatal diarrhoea kids. Frontiers in Veterinary Science, 8, 598691. doi: 10.3389/fvets.2021.598691
- Cho, Y. & Yoon, K. J. (2014). An overview of calf diarrhea-infectious etiology, diagnosis, and intervention. Journal of Veterinary Science, 15(1), 1–17. doi: 10.4142/jvs.2014.15.1.1
- Consoli, V., Sorrenti, V., Grosso, S. & Vanella, L. (2021). Heme oxygenase-1 signaling and redox homeostasis in physiopathological conditions. Biomolecules, 11(4), 589. doi: 10.3390/biom11040589
- Durán Aguilar, M., Román Ponce, S. I., Ruiz López, F. J., González Padilla, E., Vásquez Peláez, C. G., Bagnato, A. & Strillacci, M. G. (2017). Genome-wide association study for milk somatic cell score in Holstein cattle using copy number variation as markers. Journal of Animal Breeding and Genetics, 134, 49–59. doi: 10.1111/jbg.12238
- Essa, B., Al-Sharif, M., Abdo, M., Fericean, L. & Ateya, A. (2023). New insights on nucleotide sequence variants and mRNA levels of candidate genes assessing resistance/susceptibility to mastitis in Holstein and montbéliarde dairy cows. Veterinary Science, 10(1), 35. doi: 10.3390/vetsci10010035
- Fabiana, A. M., Kívia, Q. D. A., Juliana, C. F. D. S., Orlando, R. P. A. & Marília, O. F. G. (2015). Antioxidant therapy for treatment of inflammatory bowel disease: Does it work? Redox Biology, 6, 617–639. doi: 10.1016/j.redox.2015.10.006
- Feng, C., Tang, Y., Liu, X. & Zhou, Z. (2021). Cmpk2 of triploid crucian carp is involved in immune defense against bacterial infection. Developmental & Comparative Immunology, 116, 103924. doi: 10.1016/j.dci.2020.103924
- Feuk, L., Marshall, C. R., Wintle, R. F. & Scherer, S. W. (2006). Structural variants: Changing the landscape of chromosomes and design of disease studies. Human Molecular Genetics, 15, 57–66. doi: 10.1093/hmg/ddl057
- Fischer, S., Bauerfeind, R., Czerny, C. P. & Neumann, S. (2016). Serum interleukin-6 as a prognostic marker in neonatal calf diarrhea. Journal of Dairy Science, 99, 6563–6571. doi: 10.3168/jds.2015-10740
- Glasauer, A. & Chandel, N. S. (2014). Targeting antioxidants for cancer therapy. Biochemical Pharmacology, 92, 90–101. doi: 10.1016/j.bcp.2014.07.017
- Gulliksen, S. M., Jor, E., Lie, K. I., Hamnes, I. S., Løken, T., Akerstedt, J. & Osterås, O. (2009). Enteropathogens and risk factors for diarrhea in Norwegian dairy calves. Journal of Dairy Science, 92, 5057–5066. doi: 10.3168/jds.2009-2080
- Gutteridge, J. M. C. (1993). Invited review free radicals in disease processes: A compilation of cause and consequence. Free Radical Research Communications, 19, 141–158. doi: 10.3109/10715769309111598
- Hassan, S. U., Chua, E. G., Paz, E. A., Kaur, P., Tay, C. Y., Greeff, J. C., Liu, S. & Martin, G. B. (2002). Investigating the development of diarrhoea through gene expression analysis in sheep genetically resistant to gastrointestinal helminth infection. Scientific Reports, 12, 2207. doi: 10.1038/s41598-022-06001-4
- Izzo, M. M., Kirkland, P. D., Mohler, V. L., Perkins, N. R., Gunna, A. A. & House, J. K. (2011). Prevalence of major enteric pathogens in Australian dairy calves with diarrhea. Australian Veterinary Journal, 89(5), 167–173. doi: 10.1111/j.1751-0813.2011.00692.x
- Judi, H., Judi, R; & Abdul-Kareem, S. (2020). Molecular study of colibacillosis susceptibility in calves and lambs. Nano Biomedicine and Engineering, 12(2), 153–159. doi: 10.5101/nbe.v12i2.p153-159
- Kanehisa, M., Sato, Y., Furumichi, M., Morishima, K. & Tanabe, M. (2018). New approach for understanding genome variations in KEGG. Nucleic Acids Research, 47, 590–595. doi: 10.1093/nar/gky962
- Kaper, J., Nataro, J. & Mobley, H. (2004). Pathogenic Escherichia coli. Nature Reviews Microbiology, 2(2), 123–140. doi: 10.1038/nrmicro818
- Karamitros, C. S. & Konrad, M. (2014). Human 60-kDa lysophospholipase contains an N-terminal L-asparaginase domain that is allosterically regulated by L-asparagine. Journal of Biological Chemistry, 289, 12962–12975. doi: 10.1074/jbc.M113.545038
- Kirkpatrick, B. W., Cooke, M. E., Frie, M., Sporer, K. R. B., Lett, B., Wells, S. J. & Coussens, P. M. (2022). Genome-wide association analysis for susceptibility to infection by mycobacterium avium ssp. paratuberculosis in US holsteins. Journal of Dairy Science, 105, 4301–4313. doi: 10.3168/jds.2021-21276
- Klein-Jöbstl, D., Arnholdt, T., Sturmlechner, F., Iwersen, M. & Drillich, M. (2015). Results of an online questionnaire to survey calf management practices on dairy cattle breeding farms in Austria and to estimate differences in disease incidences depending on farm structure and management practices. Acta Veterinaria Scandinavica, 57, 44. doi: 10.1186/s13028-015-0134-y
- Kobayashi, A., Kang, M. I., Okawa, H., Ohtsuji, M., Zenke, Y., Chiba, T., Igarashi, K. & Yamamoto, M. (2004). Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Molecular and Cellular Biology, 24(16), 7130–7139. doi: 10.1128/MCB.24.16.7130-7139.2004
- Kukurba, K. R. & Montgomery, S. B. (2015). Rna sequencing and analysis. Cold Spring Harbor Protocols, 13(11), 951–969.
- Kumar, R., Verma, A. K., Kumar, A., Srivastava, M. & Lal, H. P. (2012). Prevalence of campylobacter spp. in Tegs attending veterinary practices at Mathura, India and risk indicators associated with shedding. Asian Journal of Animal and Veterinary Advances, 7(8), 754–760. doi: 10.3923/ajava.2012.754.760
- Li, W., Mao, L., Shu, X., Liu, R., Hao, F., Li, J., Liu, M., Yang, L., Zhang, W., Sun, M., et al. (2019). Transcriptome analysis reveals differential immune related genes expression in bovine viral diarrhea virus-2 infected goat peripheral blood mononuclear cells (PBMCs). BMC Genomics, 20, 516. doi: 10.1186/s12864-019-5830-y
- Livak, K. J. & Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT. Methods, 25, 402–408. doi: 10.1006/meth.2001.1262
- Lundgren, O. & Svensson, L. (2001). Pathogenesis of rotavirus diarrhea. Microbes and Infection, 3(13), 1145–1156. doi: 10.1016/S1286-4579(01)01475-7
- Masella, R., Di Benedetto, R., Varì, R., Filesi, C. & Giovannini, C. (2005). Novel mechanisms of natural antioxidant compounds in biological systems: Involvement of glutathione and glutathione-related enzymes. The Journal of Nutritional Biochemistry, 16, 577–586. doi: 10.1016/j.jnutbio.2005.05.013
- Mohamed, H. K., Mobasher, M. A., Ebiya, R. A., Hassen, M. T., Hagag, H. M., El-Sayed, R., Abdel-Ghany, S., Said, M. M. & Awad, N. S. (2022). Anti-inflammatory, anti-apoptotic, and antioxidant roles of honey, royal jelly, and propolis in suppressing nephrotoxicity induced by doxorubicin in male albino rats. Antioxidants, 11(5), 1029. doi: 10.3390/antiox11051029
- Morrison, S. Y., Pastor, J. J., Quintela, J. C., Holst, J. J., Hartmann, B., Drackley, J. K. & Ipharraguerre, I. R. (2017). Short communication: Promotion of glucagon-like peptide-2 secretion in dairy calves with a bioactive extract from Olea europaea. Journal of Dairy Science, 100, 1940–1945. doi: 10.3168/jds.2016-11810
- Nataro, J. P. & Kaper, J. B. (1998). Diarrheagenic Escherichia coli. Clinical Microbiology Reviews, 11, 142–201. doi: 10.1128/CMR.11.1.142
- Nathan, C. & Cunningham-Bussel, A. (2013). Beyond oxidative stress: An immunologist’s guide to reactive oxygen species. Nature Reviews Immunology, 13, 349–361. doi: 10.1038/nri3423
- Pal, A. & Chakravarty, A. K. (2020). Disease resistance for different livestock species. Genetics and Breeding for Disease Resistance of Livestock, Chapter 19, 271–296. doi: 10.1016/B978-0-12-816406-8.00019-X
- Perez, E., Noordhuizen, J. P. T. M., Van Wuijkhuise, L. A. & Stassen, E. N. (1990). Management factors related to calf morbidity and mortality rates. Livestock Production Science, 25, 79. doi: 10.1016/0301-6226(90)90043-6
- Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Research, 29, e45. doi: 10.1093/nar/29.9.e45
- Pisani, L. F., Lecchi, C., Invernizzi, G., Sartorelli, P., Savoini, G. & Ceciliani, F. (2009). In vitro modulatory effect of omega-3 polyunsaturated fatty acid (EPA and DHA) on phagocytosis and ROS production of goat neutrophils. Veterinary Immunology and Immunopathology, 131, 79–85. doi: 10.1016/j.vetimm.2009.03.018
- Radostits, O. M., Gay, C. C., Hinchcliff, K. W. & Constable, P. D. (2010). Veterinary Medicine: A Textbook of the Diseases of Cattle, Horses, Sheep, Pigs and Goats (10th ed.). Elsevier, London: Saunders, 966–994. https://www.scirp.org/(S(351jmbntvnsjt1aadkposzje))/reference/ReferencesPapers.aspx?ReferenceID=1679700.
- Ramel, S. E., Brown, L. D. & Georgieff, M. K. (2014). The impact of neonatal illness on nutritional requirements-one size does not fit all. Current Pediatrics Reports, 2(4), 248–254. doi: 10.1007/s40124-014-0059-3
- Rasko, D. A. & Sperandio, V. (2010). Anti-virulence strategies to combat bacteria-mediated disease. Nature Reviews Drug Discovery, 9, 117–128. doi: 10.1038/nrd3013
- Rosa, F. & Osorio, J. S. (2019). Short communication: Comparative gene expression analysis on the enrichment of polymorphonuclear leukocytes and gastrointestinal epithelial cells in fecal RNA from nondiarrheic neonatal dairy calves. Journal of Dairy Science, 102(8), 7464–7468. doi: 10.3168/jds.2018-16074
- Sanger, F., Nicklen, S. & Coulson, A. R. (1977). DNA sequencing with chain-terminating inhibitors. Proceedings of the National Academy of Sciences, 74, 5463–5467. doi: 10.1073/pnas.74.12.5463
- Schaeffer, L. R. (2006). Strategy for applying genome-wide selection in dairy cattle. Journal of Animal Breeding and Genetics, 123, 218–223. doi: 10.1111/j.1439-0388.2006.00595.x
- Semrad, C. E. (2012). Approach to the patient with diarrhea and malabsorption. Goldman's Cecil Medicine, 1, 895–913. doi: 10.1016/B978-1-4377-1604-7.00142-1
- Shi, H. R., Huang, X. Y., Yan, Z. Q., Yang, Q. L., Wang, P. F., Li, S. G., Sun, W. & Gun, S. (2019). Effect of clostridium perfringens type C on TLR4/MyD88/NF-κB signaling pathway in piglet small intestines. Microbial Pathogenesis, 135, 7. doi: 10.1016/j.micpath.2019.103567
- Sivula, N. J., Ames, T. R. & Marsh, W. E. (1996). Management practices and risk factors for morbidity and mortality in Minnesota dairy heifer calves. Preventive Veterinary Medicine, 27, 173–182. doi: 10.1016/0167-5877(95)01001-7
- Spees, A. M., Wangdi, T., Lopez, C. A., Kingsbury, D. D., Xavier, M. N., Winter, S. E., Tsolis, R. M. & Bäumler, A. J. (2013). Streptomycin-induced inflammation enhances Escherichia coli gut colonization through nitrate respiration. Mbio, 4, e00430-13. doi: 10.1128/mBio.00430-13
- Tamura, K., Dudley, J., Nei, M. & Kumar, S. (2007). MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Molecular Biology and Evolution, 24, 1596–1599. doi: 10.1093/molbev/msm092
- Tang, F., Lao, K. & Surani, M. A. (2011). Development and applications of single-cell transcriptome analysis. Nature Methods, 8(4), 6–11. doi: 10.1038/nmeth.1557
- Tsuchida, S., Yamad, Y., Fukui, E., Kawada, T., Omi, T., Tsuchida, A., Sako, T., Hatakeyama, H. & Kotani, K. (2010). Distribution of single nucleotide polymorphisms in the CXCR1 gene and association with calf diseases in Japanese black cattle. Journal of Veterinary Medical Science, 72, 1609–1614. doi: 10.1292/jvms.10-0050
- van Harten, S., Brito, R., Almeida, A. M., Scanlon, T., Kilminster, T., Milton, J., Greeff, J., Oldham, C. & Cardoso, L. A. (2013). Gene expression of regulatory enzymes involved in the intermediate metabolism of sheep subjected to feed restriction. Animal, 7(3), 439–445. doi: 10.1017/S1751731112001589
- von Buenau, R., Jaekel, L., Schubotz, E., Schwarz, S., Stroff, T. & Krueger, M. (2005). Escherichia coli strain Nissle 1917: Significant reduction of neonatal calf diarrhea. Journal of Dairy Science, 88, 317–323. doi: 10.3168/jds.S0022-0302(05)72690-4
- Wadleya, A. J., Aldred, S. & Coles, S. (2016). An unexplored role for Peroxiredoxin in exercise-induced redox signalling? Redox Biology, 8, 51–58. doi: 10.1016/j.redox.2015.10.003
- Warnick, L. D., Erb, H. N. & White, M. E. (1994). The association of calfhood morbidity with first calving age and dystocia in New York Holstein herds. Kenya Veterinary, 18, 177.
- Wei, W., Feng, W., Xin, G., Niu, T. & Yan, X. (2016). Enhanced effect of κ-carrageenan on TNBS-induced inflammation in mice. International Immunopharmacology, 39, 218–228. doi: 10.1016/j.intimp.2016.07.031
- Wiltshire, S. A., Leiva-Torres, G. A. & Vidal, S. M. (2011). Quantitative trait locus analysis, pathway analysis, and consomic mapping show genetic variants of TNNI3K, FPGT, or H28 control susceptibility to viral myocarditis. The Journal of Immunology, 186, 6398–6405. doi: 10.4049/jimmunol.1100159
- Wu, H. J. & Wu, E. (2012). The role of gut microbiota in immune homeostasis and autoimmunity. Gut Microbes, 3(1), 4–14. doi: 10.4161/gmic.19320
- Yang, Q. L., Zhao, S. G., Wang, D. W., Feng, Y., Jiang, T. T., Huang, X. Y. & Gun, S. B. (2014). Association between genetic polymorphism in the swine leukocyte antigen-DRA gene and piglet diarrhea in three Chinese pig breeds. Asian-Australasian Journal of Animal Sciences, 27, 1228–1235. doi: 10.5713/ajas.2013.13567
- Younis, E. E., Ahmed, A. M., El-Khodery, S. A., Osman, S. A. & El-Naker, Y. F. (2009). Molecular screening and risk factors of enterotoxigenic Escherichia coli and Salmonella spp. in diarrheic neonatal calves in Egypt. Research in Veterinary Science, 87, 373–379. doi: 10.1016/j.rvsc.2009.04.006
- Zaki, E. R. (2003). Evaluation of effeminacies of some vaccine used for production of new born Buffalo calves against E.coli k 99 by ELISA. Journal of the Egyptian Veterinary Medical Association, 63, 275–284.
- Zhang, X., Zhang, K. & Zhang, Y. (2020). Pigment epithelium–derived factor facilitates NLRP3 inflammasome activation through downregulating cytidine monophosphate kinase 2: A potential treatment strategy for missed abortion. International Journal of Molecular Medicine, 45, 1436–1446.