ABSTRACT
The experimental system: Sweetpotato (Ipomoea batatas [L.] Lam.) is an important food crop widely grown under low input production systems and harsh growing environments. It is a relatively drought tolerant crop attaining higher biomass production per unit area. Genetic diversity present in breeding populations is a raw material for selection of parental genotypes with desirable and complementary traits. The objective of this study was to determine the genetic diversity present among Tanzania grown sweetpotato germplasm to select promising breeding parents with enhanced yield and yield-related traits and dry matter content. Procedures: Seventy six sweetpotato accessions collected from Tanzania and 20 sweetpotato accessions received from International Potato Centre (CIP) in Lima/Peru were characterized in two seasons. The study was conducted using a 16 x 6 triple lattice design. The data collected included 16 morphological traits using CIPs standard descriptors. Data were analysed using multivariate procedure including cluster analysis and principle component analysis. Results and conclusions: The tested sweetpotato collections differed significantly for storage root yield, dry matter content (DMC) and number of roots per plot. Genotypes New Kawogo, Kiti cha Nyerere and Kisu cha Masai had the highest root yields whereas genotypes Ngw’anangusa, Rugomoka and Secondary had significantly higher mean DMC. Traits considered in the study revealed positive and significant correlations. The first four principal components accounted for 69.33% of the variations present in the tested sweetpotato genotypes. Cluster analysis grouped the studied genotypes into two major classes with genetic diversity of 0.54. The selected genotypes can be recommended for future breeding programs to bolster yield and dry matter content of sweetpotato under western Tanzania conditions.
Introduction
Sweetpotato (Ipomoea batatas [L.] Lam.) is the world’s seventh most important food crop (Elameen et al. Citation2008; Nelles Citation2009). In sub-Saharan African (SSA) countries sweetpotato is cultivated as a food security crop. It is a highly preferred crop by smallholder farmers owing to its ability to grow under marginal soil and dryland environmental conditions yet attaining higher yield per unit area compared to wheat, rice and cassava (Schafleitner et al. Citation2010). The crop is early maturing (3 to 4 months) hence able to escape drought under short rainfall condition.
The storage root of sweetpotato is consumed as fried chips, boiled root or as baked products (Engoru et al. Citation2005). Also the root is an industrial raw material to extract starch, alcohol, biofuel or for animal feed (Schafleitner et al. Citation2010; Clark et al. Citation2012). Young and succulent leaves of the crop are used as leaf vegetable as well as fodder crop. The orange fleshed sweetpotato (OFSP) varieties are rich in carbohydrate and β-carotene content that is a precursor of vitamin ‘A’ useful in combating vitamin A deficiency (Mwanga et al. Citation2007; Burri Citation2011).
Tanzania is amongst the top 5 leading world producers of sweetpotato (FAOSTAT Citation2014). In the country sweetpotato is cultivated on 560 000 ha of agricultural lands with a mean national yield of 4.55 t ha−1 (FAOSTAT Citation2014). This yield level is far below the potential productivity of the crop varying from 15 to 23 t ha−1 reached under 3 0 kg N ha−1 and 60 kg P ha−1 level of fertilization (Sebastiani et al. Citation2007). Moreover there is limited progress in sweetpotato breeding in Tanzania due to several constraints that include lack of knowledge on the genetic diversity of the crop among locally grown genotypes (Kapinga and Carey Citation2003; Tairo et al. Citation2008). Often locally grown genotypes bear different names despite their considerable genetic similarity. A report by Kapinga et al. (Citation1995) revealed that there were more than 100 local names for sweetpotato varieties grown in the Maswa district in Tanzania. In some cases, the same variety bear different names in different location and/or different varieties may bear the same name. Therefore, there is need to systematically characterize and select genotypes with high yields and high dry matter content for effective breeding of the crop.
Genetic diversity analysis of germplasm collections can be undertaken using agro-morphological traits and molecular markers. Some of the molecular markers used in genetic analysis of sweetpotato included simple sequence repeat (SSR), random amplified polymorphic DNA (RAPD), amplified fragment length polymorphism (AFLP), restriction fragment length polymorphism (RFLP), allozyme and single nucleotide polymorphism (SNP) (Acquaah Citation2012). Morphological markers have been widely and successively used in studying the diversity of sweetpotato genotypes (Tairo et al. Citation2008; Vimala and Hariprakash Citation2011; Mbithe et al. Citation2016; Shumbusha et al. Citation2017).
Genetic diversity present in a breeding population is vital for selecting promising genotypes with desirable and complementary traits. Different studies reported the availability of a considerable level of genetic diversity of the crop in Tanzania. Elameen et al. (Citation2008) used amplified fragment length polymorphism (AFLP) and studied the genetic diversity of 97 accessions of the crop from Eastern Tanzania and reported an average genetic similarity of 0.71. Tairo et al. (Citation2008) examined genetic diversity of the crop using 136 landraces collected from Lake Zone, eastern and southern Tanzania and reported a genetic diversity of 0.52. Gwandu et al (Citation2012.) used 57 sweetpotato genotypes collected from eastern and southern Tanzania and subjected for single sequence repeat (SSR) analysis to investigate the genetic diversity of the crop for sweetpotato virus disease (SPVD) resistance and dry mass content. The authors reported a mean pair-wise genetic distance of 0.55. Ngailo et al. (Citation2016) used SSR markers and characterized 48 sweetpotato genotypes collected from Lake and eastern zones of Tanzania. This study reported a mean number of alleles amplified per locus at 9.78. The above studies indicated the presence of a considerably higher level of genetic diversity among Tanzanian sweetpotato genotypes. Previous studies used sweetpotato germplasm collections from eastern, southern and lake zones of Tanzania. Thus far there is no report on the genetic diversity of the crop using germplasm collections from western Tanzania. The objective of this study was to determine the genetic diversity present among sweetpotato germplasm widely grown in western Tanzania using morphological markers to select promising parents with enhanced yield and yield-related traits and dry matter content.
Materials and methods
Description of the study site
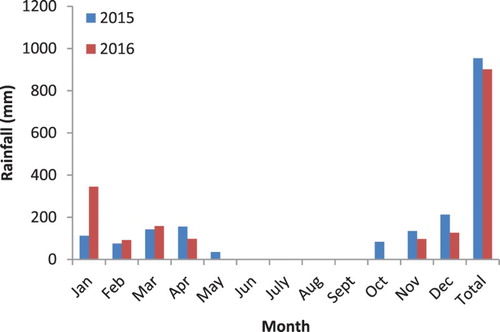
The study was conducted at Tumbi Agricultural Research Institute (ARI-Tumbi) situated at 5°4′11″S, 32°40′1″E in western Tanzania. The area is characterized by a unimodal rainfall pattern with a mean annual rainfall of 920 mm. The main rainfall season is between November and April followed by 6 months dry season. The rainfall distribution at the study site during the two seasons of the study is presented in . The climate is generally dry and warm with a mean daily temperature of 23°C. The soils are slightly acidic predominantly with 67% sand, 24% clay and 9% silt (Bagarama et al. Citation2012).
Germplasm collection and trial establishment
A total of 82 sweetpotato genotypes were collected during October and November 2014 from seven districts of western Tanzania and Lake Zone while 20 sweetpotato genotypes were received from CIP-Peru. The collected genotypes were multiplied at ARI-Tumbi. During multiplication some duplicate genotypes from western Tanzania and Lake Zone were excluded based on their morphological similarities leaving 96 germplasm genotypes (). Among the 96 genotypes, 57 were local varieties or landraces collected from western Tanzania; 19 were collected from Lake Zone and Maruku and Ukiriguru Research Institutes. Twenty genotypes were kindly supplied by International potato centre (CIP)–Lima/Peru. The CIP collections were originally obtained from India. Trials were conducted at ARI-Tumbi from February to June 2015 (season I) and January to May 2016 (season II). The experiment was designed using a 16 × 6 lattice with 3 replications. The plot size was 1.3 square metres (1 × 1.3 m) composed of two ridges. Five plants were established on each ridge making a total of 10 plants per plot. The spacing used was 1 m inter-row and 0.3 m intra-row. Weeding was done twice using hand hoe. Fertilizers were applied using Nitrogen, Phosphorus and Potassium (23:10:5) at the rate of 233 kg/ha a month after planting. This fertilization rate was used to boost vegetative growth because the soils at trial site (ARI Tumbi) are dominated by sand (Bagarama et al. Citation2012).
Table 1. List, origin, and description of sweetpotato genotypes used in the study.
Data collection
Test genotypes were phenotyped using 16 morphological traits. Characterization was done using CIP’s standard descriptors (Huaman Citation1991). Data were averaged from measurements made on five plants of each accession. The colour and pigmentations on leaves and vines were recorded as the average expression of the character observed in a section of the main stem located in the middle portion of 5 main stems. The detailed scores on descriptors used are presented in Supplemental Table 1. Dry matter content was determined by taking a sample of 100–200 g fresh storage root mass from five randomly selected healthy roots of each genotype followed by chopping roots into smaller sections. Each sliced root sample was oven dried at 70°C for 72 h to constant mass. The dry matter content was determined as the proportion of dry matter relative to fresh mass expressed as a percentage.
Data analysis
Restricted maximum likelihood (REML) procedure (Payne et al. Citation2015) was used to analyse the variations of genotypes for yield and yield components. Genotype was treated as fixed effect whereas season and genotype by season interaction, replication and block were treated as random effects. The model for REML analysis was as follows:Where: μ is the general mean, G is the genotype effects, S is the season effects, GS is the interaction effects of genotype and season, R is replication effects, B is the block effects and ϵ is the random term. The mean separation was done using Fisher’s unprotected least significant difference (LSD) at P ≤ 0.05.
Pearson’s correlation coefficients were calculated using SPSS 20 programme (SPSS Citation2009) to assess the relationship between traits. Cluster analysis by Ward’s method was done using the squared Euclidean distance to classify the genotypes into main groups and sub-groups.
Principle component analysis (PCA) was conducted using GENSTAT 14th edition programme (Payne et al. Citation2015) to detect which descriptor variables contributed more to the variations among sweetpotato genotypes. The latent vectors were used to select the principle components which explained more of the variations among the tested sweetpotato genotypes. The vector loadings were used to ascertain which descriptor variables highly correlated with principle components which explain more of the variations among sweetpotato genotypes. Hierarchical cluster analysis based on complete linkage method was used to classify genotypes into main groups and sub-groups and genetic similarity matrix using 16 key morphological descriptors.
Results
Storage root number, root yield and dry matter content
There was a significant difference (P < 0.001) among genotypes for number of roots per plot, root yield and dry matter content (). Season had a significant effect (P < 0.001) on root number per plot and yield t ha−1. There was also a significant effect of variety by season interaction for these traits. The mean root number per plot (1.3 square metres) ranged from 4.50 to 36.20. The genotypes Jewel, 18-CIP and Kisu cha Masai had significantly higher mean root number of 36.20, 32.20 and 31.20, in that order (). The storage root yield ranged from 1.72 to 10.14 t ha−1. Genotype New Kawogo collected from Lake Zone had the highest yield of 10.14 t ha−1 followed by Kiti cha Nyerere and Kisu cha Masai all collected from western Tanzania and yielding 9.85 and 9.67 t ha−1 respectively. The following genotypes: Kibandule, Madebe and Kabelele had significantly low mean root yields of 1.72, 2.65 and 2.85 t ha−1 respectively. Genotypes expressed high yields during season I than season II. This could be attributed to a relatively better rainfall distribution in season I (). DMC varied from 27.40% to 43.50% (). The following genotypes: Ngw’anangusa and Rugomoka collected from Western Zone and genotype Secondary collected from Lake Zone had significantly higher dry matter content of 43.50%, 43.30% and 43.30% respectively (). Genotypes 20-CIP, Chuga and Awilo had significantly lower DMC of 27.40%, 31.40% and 31.50%, respectively.
Table 2. Wald statistic showing significant tests for root yield, dry matter content and number of roots per plant.
Table 3. Mean root number, root yield, and dry matter content of sweetpotato genotypes assessed in two seasons in western Tanzania.
Correlation among traits
The correlations among quantitative traits are presented in (). There was a significant positive correlation (P < 0.01) between root yield and root number per plot, dry matter content and ground cover, root yield and ground cover, (). A significant negative correlation (−0.21) was revealed between root number and dry matter content.
Table 4. Pearson correlation coefficients showing pair-wise association of 11 quantitative traits among 96 sweetpotato genotypes evaluated in western Tanzania.
Principal component analysis (PCA)
The first four principal components with latent roots value > 1 accounted for 69.33% of the variations among sweetpotato genotypes. PC1 was highly and positively correlated (0.90) with the root number per plot, and root yield (0.20) but highly and negatively correlated with root dry matter content (0.14). PC2 was highly and positively correlated (0.56) with root dry matter content, while PC3 was highly and positively correlated (0.47) with root dry matter content and root number per plot (0.20). PC4 was highly and positively correlated (0.56) with root dry matter content.
Clustering of sweetpotato genotypes based on phenotypic traits
The cluster analysis resulted in two major groups: Group A and Group B (). Group A had 11 sub-groups consisting of 66 genotypes whereas Group B had 5 sub-groups with 30 genotypes. The analysis showed a genetic diversity of 0.54 among test genotypes. Clustering of sweetpotato genotypes in groups showed a lack of association between origin of collection and genotypes. However most of the test genotypes from all the three geographical areas were clustered in Group A. Sub-group B3 had 4 genotypes all obtained from CIP/Peru ().
Table 5. Main genetic groups, sub-groups and name of genotypes based on cluster analysis using complete linkage method.
Genetic similarities between and within groups and sub-groups of characterized sweetpotato genotypes ranged from 0.66 to 0.91 with the mean similarity value of 0.85. The highest similarity of 0.91 was expressed within sub-group A2 consisting of 4 genotypes collected from Lake Zone, 3 from CIP and 4 from western Tanzania. The lowest similarity of 0.66 was expressed within sub-group B5 (consisting of 2 genotypes from Lake Zone and 1 from western Tanzania) and sub-group A10 (with 3 genotypes from western Tanzania and 1 from CIP.
Discussion
Storage root yield
Sweetpotato genotypes used in this study were collected from different agro-ecological areas () hence had differences in adaptability to the trial site leading to significant difference in root yield (). Sweetpotato varieties differ in their interaction with the environment in which they are growing. Varieties tend to have high yield in the environment in which they are well-adapted (Acquaah Citation2012). The variation in root yield among tested sweetpotato genotypes also might be attributed to genetic differences among test entries (Kapinga et al. Citation2003; Lebot Citation2009; Acquaah Citation2012; Rukundo et al. Citation2013). Sweetpotato varieties differ in their genetic make-ups controlling accumulation of dry matter, growth and development of storage roots (Rukundo et al. Citation2013). Lebot (Citation2009) suggested that root yield is the function of growing time. The more the time a crop takes to grow under field condition the more dry matter it accumulates and thus more yield. Therefore the difference in time to maturity among sweetpotato varieties also might have contributed to their differences in root yield. Significant difference in root yield has been found among sweetpotato varieties in other studies (Tairo et al. Citation2008; Ngailo et al. Citation2016). Season one also had higher root yield than season two due to difference in rainfall distribution among the two seasons (). In this study, genotypes New Kawogo, Kiti cha Nyerere and Kisu cha Masai had significantly higher yield potential (). These genotypes are promising parents to be used in sweetpotato breeding programmes to enhance yield and yield related traits.
Dry matter content and root number
Dry matter content is an important criterion for sweetpotato variety selection. For instance, in sub-Saharan Africa farmers prefer sweetpotato genotypes with high dry matter content (> 28%), moderate sweetness and with dry mouth feel, while in the continental America farmers prefer sweetpotato varieties with low dry matter content, moist mouth feel and very sweet (Grüneberg et al. Citation2009; Cervantes-Flores et al. Citation2011). Dry matter content also is a crucial quality trait for sweetpotato cultivar adoption (Grüneberg et al. Citation2009; Cervantes-Flores et al. Citation2011).
Gasura et al (Citation2010.) observed marked differences among genotypes in dry matter content and root number per plot. The significant difference in dry matter content and root number may be attributed to genotypic variations. For instance the expression of storage root formation and dry matter accumulation are controlled by different genes which differ among varieties (Rukundo et al. Citation2013). Dry matter accumulation in storage root also depends on the ability of the plant to translocate the photosynthetic assimilates from shoots to underground roots. Dry matter content decreases as the number of storage roots increases because it becomes difficult for a plant to supply enough photosynthetic assimilates to all the roots (Gasura et al. Citation2010). The presently tested genotypes Ngw’anangusa, Rugomoka and Secondary were found to have the highest dry matter content (). These are selected as the best parent for breeding to enhance high dry matter content.
Correlation among traits
Most traits considered in this study revealed a significant positive correlation (). For instance a significant correlation (0.74) was revealed between root yield and root number. Other studies have also reported a significant positive correlation between root number and root yield (Gasura et al. Citation2010; Ngailo et al. Citation2016). Also, significant correlations were calculated between dry matter content and ground cover (0.48), root yield and ground cover (0.32). Selection of correlated traits influences each other thus allowing simultaneous selection in plant breeding programmes (Acquaah Citation2012; Rukundo et al. Citation2013). In the current study a negative correlation (−0.21) was also revealed between root number and dry matter content (). This implies that selection for high dry matter content reduces the number of roots per plant. This result conforms to the previous findings that dry matter content decreases as the number of storage roots increases because it becomes difficult for a plant to supply enough photosynthetic assimilates to all the roots (Gasura et al. Citation2010).
Genetic diversity
The current study anticipated higher genetic diversity among test genotypes given the inclusion of some germplasm from CIP. However, minimal genetic diversity was detected. The CIP genotypes used in this study, though introduced from Peru, had their origins from India. Both Tanzania and India are neither the centre of diversity nor centre of origin of sweetpotato. The present study found a genetic diversity of 0.54 when using CIP standard morphological descriptors of sweetpotato geneotypes. This value is at par with the value of 0.52 found among sweetpotato genotypes collections and evaluations made from 3 different agro-ecological zones of Tanzania (Tairo et al. Citation2008). Genetic diversity of 0.55 was reported using simple sequence repeat markers (Gwandu et al. Citation2012). Therefore the observed low genetic diversity among the characterized germplasm in this study might be due to the fact that both geographical areas received sweetpotato variety introductions from the same genetic pool.
The current results showed higher genetic similarity of 0.85 when genotypes were evaluated using agro-morphological traits. This value is higher than the value of 0.71 found in Tanzania sweetpotato germplasm using the amplified fragment length polymorphism (AFLP) (Elameen et al. Citation2008). Sub-group A2 expressed the highest genetic similarity of 0.91 due to varied sources of genotypes from all geographical sources (i.e. 4 from Lake Zone, 3 from CIP and 4 from Western Tanzania). The lowest genetic similarity was expressed in sub-group B5 and sub-group A10. Sub group B5 had 2 genotypes from Lake Zone and 1 from western Tanzania, while sub-group A10 had 3 genotypes from western Tanzania and 1 from CIP. This result suggests that there are more variations within the region than between regions. The high genetic similarity revealed in this study among genotypes of different geographical sources is in agreement with the result reported by Elameen et al (Citation2008) and it is attributed to common genetic pool of the genotypes characterized. The current results also conform to earlier findings that sweetpotato germplasm express more variations within the geographic region than between geographical regions (Gichuki et al. Citation2003). The authors proposed that this may be due to natural mutational events, inbreeding and some introductions of new genotypes.
Clustering of sweetpotato genotypes
Cluster analysis grouped the tested sweetpotato genotypes into two main groups, group A and group B (). The same trend of grouping was reported by other researchers characterizing Tanzanian sweetpotato genotypes (Elameen et al. Citation2008; Tairo et al. Citation2008; Gwandu et al. Citation2012).These reports indicated that the two main grouping of sweetpotato could be resulted due to introductions of sweetpotato to Tanzania. It is believed that the crop was introduced into the country firstly by Portuguese and secondly by British (Kapinga et al. Citation1995). Elameen et al. (Citation2008) suggested that the two main introductions of sweetpotato to Tanzania might have come from two different genetic pools. The two main groups formed by cluster analysis in this study () can serve as important genetic clusters for breeding.
Two main groups of genotypes presented in by cluster analysis, were formed in a random manner irrespective of their geographical origin. Proper naming of sweetpotato varieties is the first and easiest criteria in distinguishing sweetpotato varieties. However farmers in Tanzania use unstandardized system of naming leading to the same variety bearing different names in different areas (Kapinga et al. Citation1995; Tairo et al. Citation2008). For instance in the current study, Simama is an improved variety which is slightly orange fleshed, the same variety is called Mayai in Nzega district and Kayai in Sikonge district. This way the same variety can be considered bearing three different varieties. However, the present study could not detect any phenotypic difference in shoot and root morphology confirming these entries as duplicates. In some cases different varieties bear the same name but are basically different though the difference can be less conspicuous. For instance in Kigoma region three different varieties bear the same name of Masinia. In this situation one variety can be sampled, while excluding the other two. However, some farmers cautioned and suggested that though the varieties bear the same name they are different. Therefore the present study named the three genotypes as Masinia Mguu wa kware, Masinia njano and Masinia nyeupe. These collections had phenotypic resemblance in their shoot morphology and skin colour. However they have some slight differences in flesh colour and root latex production.
In summary the present study detected a genetic diversity of 0.54 in the sampled sweetpotato germplasm. The cluster analysis also grouped the sweetpotato germplasm into two main groups. The genetic diversity detected in the study and the two main genetic groups of germplasm can be exploited by sweetpotato breeders to breed new varieties. Genotypes Ngw’anangusa, Rugomoka and Secondary were selected with the highest dry matter content, while genotypes New Kawogo, Kiti cha Nyerere and Kisu cha Masai had the highest storage root yield. These genotypes are recommended as the best parents for sweetpotato breeding to enhance yield and dry matter content in western Tanzania or similar agro-ecologies.
Acknowledgements
Gratitude is due to the International Potato Center (CIP) and Ukiriguru Agricultural Research Institute (ARI Ukiriguru) for sharing sweetpotato germplasm. The first author is grateful to the Tanzanian Ministry of Agriculture, Livestock and Fisheries through Tumbi Agricultural Research Institute (ARI-Tumbi) for providing leave of absence and research support.
Disclosure statement
No potential conflict of interest was reported by the authors.
Notes on contributors
Mr. Filson Mbezi Kagimbo is currently working with Tanzania Ministry of Agriculture, Livestock and Fisheries at a position of “Senior Agricultural Research Officer” since 2006 to present. He is under the department of Research and Development in Root and tuber crops sub-program. His main duties include generation and dissemination of different agricultural technologies to agricultural stakeholders; conducting seminars and other training to agricultural stake holders; conducting agricultural research work, data collection, analysis and technical report writing. Mr. Kagimbo is currently undertaking his postgraduate studies (PhD) in Plant breeding in his final year at the University of KwaZulu-Natal in South Africa. The course started in January 2014 and the main focus is breeding sweetpotato varieties with resistance to sweetpotato weevils. This course is funded by the Alliance for a Green Revolution in Africa (AGRA) through African Centre for Crop Improvement. Previously, Mr. Kagimbo graduated with MSc. in Agricultural Science in 2011 at the University of Melbourne in Australia. Mr. Kagimbo also graduated with BSc. in Agronomy in 2005 at Sokoine University of Agriculture.
Professor Hussein Shimelis (Ph.D.) is a Crop Scientist with a specialization in Plant Breeding. He is Chair of Crop Science, Associate Professor of Plant Breeding and Deputy Director of the African Center for Crop Improvement (ACCI) at the University of KwaZulu-Natal, South Africa.
Dr Julia Sibiya is a Senior Lecturer in Plant Breeding, SAEES, University of KwaZulu-Natal (UKZN) in Pietermaritzburg (from 2015 to current). She received her BSc. Honours degree in Crop Science from the University of Zimbabwe, MSc in Plant Pathology from the Ohio State University, USA and PhD degree in Plant Breeding from the University of KwaZulu-Natal (UKZN), South Africa. Dr Sibiya spent 10 years as a lecturer in Plant Pathology at the University of Zimbabwe before joining UKZN’s African Centre for Crop Improvement (ACCI) in 2005 as a PhD student (Plant Breeding), and then as a lecturer from 2011 to July 2015. Dr Sibiya lectures modules in Plant Breeding including Biotechnology in Crop Improvement and Plant Breeding Design and Management. She also supervises PhD students from different African countries in the ACCI. Her research focus is on breeding for biotic stresses such as diseases in various crops, particularly foliar diseases of maize and development of unique maize source germplasm, as well as sterile sorghum lines to be used in breeding for sorghum hybrids. Dr Sibiya is also the project manager of a MSc in Plant Breeding for Africa project (Improved Masters in Cultivar Development in Africa – IMCDA) funded by the Alliance for a Green Revolution in Africa (AGRA). The MSc program focuses on training industry-ready, middle-level, graduates from the Southern African Development Community (SADC) and is a joint project with Iowa State University (ISU) (curriculum development) and African partner universities – Makerere University and Kwame Nkrumah University of Science and Technology responsible for training for East and West Africa, respectively. She is also the local coordinator of the Intra Africa Academic Mobility – Mobreed project funded by the EU which focuses on training PhD and MSc students in breeding of orphan or under-utilized crops.
Additional information
Funding
References
- Acquaah G. 2012. Principles of plant genetics and breeding. 2nd ed. Oxford (USA): Wiley-Blackwell.
- Bagarama FM, Shenkalwa EM, Matata ZP. 2012. The effect of gypsum and NPK fertiliser on groundnut performance in Western Tanzania. Third RUFORUM Biennial Meeting.
- Burri BJ. 2011. Evaluating sweet potato as an intervention food to prevent vitamin A deficiency. Compr Rev Food Sci Food Safety. 10:118–130. doi: 10.1111/j.1541-4337.2010.00146.x
- Cervantes-Flores JC, Sosinski B, Pecota KV, Mwanga ROM, Catignani GL, Truong VD, Watkins RH, Ulmer MR, Yencho GC. 2011. Identification of quantitative trait loci for dry-matter, starch, and β-carotene content in sweetpotato. Mol Breed. 28:201–216. doi: 10.1007/s11032-010-9474-5
- Clark CA, Davis JA, Abad JA, Cuellar WJ, Fuentes S, Kreuze JF, Gibson RW, Mukasa SB, Tugume AK, Tairo FD, et al. 2012. Sweetpotato viruses: 15 years of progress on understanding and managing complex diseases. Plant Dis. 96:168–185. doi: 10.1094/PDIS-07-11-0550
- Elameen A, Fjellheim S, Larsen A, Rognli OA, Sundheim L, Msolla S, Masumba E, Mtunda K, Klemsdal SS. 2008. Analysis of genetic diversity in a sweet potato (Ipomoea batatas L.) germplasm collection from Tanzania as revealed by AFLP. Genet Resour Crop Evol. 55:397–408. doi: 10.1007/s10722-007-9247-0
- Engoru P, Mugisha J, Bashaasha B. 2005. Tuber utilisation options among sweet potato producers in eastern Uganda. African Crop Science Conference Proceedings.
- FAOSTAT. 2014. Food and agriculture organisation of the United Nations: crop production data. Rome: FAOSTAT Division.
- Gasura E, Mashingaidze A, Mukasa S. 2010. Genetic variability for tuber yield, quality, and virus disease complex traits in Uganda sweetpotato germplasm. Afr Crop Sci J. 16:147–160. doi: 10.4314/acsj.v16i2.54355
- Gichuki ST, Berenyi M, Zhang D, Hermann M, Schmidt J, Glössl J, Burg K. 2003. Genetic diversity in sweetpotato [Ipomoea batatas (L.) Lam.] in relationship to geographic sources as assessed with RAPD markers. Genet Resour Crop Evol. 50:429–437. doi: 10.1023/A:1023998522845
- Grüneberg W, Mwanga R, Andrade M, Espinoza J, Ceccarelli S, Guimarães E, Weltzien E. 2009. Selection methods. Part 5: breeding clonally propagated crops. Plant Breed Farmer Part. 13:275–322.
- Gwandu C, Tairo F, Mneney E, Kullaya A. 2012. Characterization of Tanzanian elite sweet potato genotypes for sweet potato virus disease (SPVD) resistance and high dry matter content using simple sequence repeat (SSR) markers. Afr J Biotechnol. 11:9582–9590.
- Huaman Z. 1991. Descriptors for the characterization and evaluation of sweet potato genetic resources. CIP Lima, Perú; p. 331–355.
- Kapinga R, Carey E. 2003. Present status of sweetpotato breeding for eastern and southern Africa. Sweetpotato post-harvest assessment: experiences from East Africa. Chatham, United Kingdom. NRI/CPHP/DFID/CIP/Ministry of Agriculture. Natural Resources Institute: The University of Greenwich. ISBN 0 859554: 2.
- Kapinga R, Jeremiah S, Rwiza E, Rees D. 2003. Farmers criteria for selection of sweetpotato varieties, results from Tanzania. Chatham (UK): Natural Resources Institute, University of Greenwich.
- Kapinga RE, Ewell PT, Jeremiah S, Kileo R. 1995. Sweetpotato in Tanzanian farming and food systems: implications for research. Nairobi: International Potato Center, Tanzanian Ministry of Agriculture.
- Lebot V. 2009. Tropical root and tuber crops: cassava, sweet potato, yams and aroids. Wallingford: CABI.
- Mbithe MJ, Steven R, Agili S, Kivuva MB, Kioko WF, Kuria E. 2016. Morphological characterisation of selected Ugandan sweet potato (Ipomoea batatas L) varieties for food and feed. Phylogen Evol Biol. 4. doi:10.4172/2329-9002.1000163.
- Mwanga ROM, Odongo B, Niringiye C, Kapinga R, Tumwegamire S, Abidin PE, Carey EE, Lemaga B, Nsumba J, Zhang D. 2007. Sweetpotato selection releases: lessons learnt from Uganda. Afr Crop Sci J. 15:11–23.
- Nelles W. 2009. Sweetpotato education, research and capacity development through a CIP-Orissa learning site. Bhubaneswar (OR): CTCRI.
- Ngailo S, Shimelis H, Sibiya J, Mtunda K. 2016. Screening of Tanzanian sweet potato germplasm for yield and related traits and resistance to sweet potato virus disease. Acta Agric Scand Sect B Soil Plant Sci. 66:52–66. doi:10.1080/09064710.2015.1063684.
- Payne R, Welham S, Harding S. 2015. A guide to REML in genstat. VSN International, 2 Amberside, Wood Lane, Hemel Hempstead, Hertfordshire HP2 4TP, UK.
- Rukundo P, Shimelis H, Laing M, Gahakwa D. 2013. Storage root formation, dry matter synthesis, accumulation and genetics in sweet potato. Aust J Crop Sci. 7:2054–2061.
- Schafleitner R, Tincopa LR, Palomino O, Rossel G, Robles RF, Alagon R, Rivera C, Quispe C, Rojas L, Pacheco JA, et al. 2010. A sweetpotato gene index established by de novo assembly of pyrosequencing and Sanger sequences and mining for gene-based microsatellite markers. BMC Genomics. 11:604–614. doi: 10.1186/1471-2164-11-604
- Sebastiani S, Mgonja A, Urio F, Ndondi T. 2007. Agronomic and economic benefits of sweetpotato (Ipomoea batatas) response to application of nitrogen and phosphorus fertilizer in the Northern highlands of Tanzania. 8th African Crop Science Society Conference, El-Minia, Egypt; October 27–31, 2007; African Crop Science Society.
- Shumbusha D, Shimelis H, Laing M, Asiimwe T. 2017. Phenotypic diversity analysis of sweetpotato for breeding dual-purpose varieties. Acta Agric Scand Sect B Soil Plant Sci. 67:340–351.
- SPSS. 2009. Statistical package for social scientists. SPSS for Windows Release 18.0, Chicago (IL).
- Tairo F, Mneney E, Kullaya A. 2008. Morphological and agronomical characterization of sweet potato [Ipomoea batatas (L.) Lam.] germplasm collection from Tanzania. Afr J Plant Sci. 2:77–85.
- Vimala B, Hariprakash B. 2011. Variability of morphological characters and dry matter content in the hybrid progenies of sweet potato (Ipomoea batatas (L) Lam). Gene Conserve. 10:65–86.