Abstract
A rapid protocol for polar lipid profiling was applied to Euglena gracilis lipid metabolism by LipidBlast, an MS/MS spectral similarity search tool. The similarity search results suggested anoxia-induced polar lipid metabolism in Euglena characterized by the accumulation of differential lipid classes, carbon chain lengths, and unsaturated bond numbers. The informatics-supported MS spectral search provides an alternative option for global lipid profiling studies.
Global profiling of polar lipids constitutes a core technology in a variety of applied research areas. Liquid chromatography–mass spectrometry (LC–MS) analysis with mass spectral library search is exemplary,Citation1,Citation2) but both the identification and the structural estimation of lipid species are hampered by the limited availability of authentic standard compounds even when used in combination with high-resolution tandem mass spectrometry, such as Fourier transform ion cyclotron resonance mass spectrometry (FT-ICR/MS) and Orbitrap-MS. For example, the MS/MS spectra of polar lipids are very limited even in MassBank (http://www.massbank.jp/), the largest publicly available database, which stores more than 15,000 MS/MS spectra for 4,000 compounds.Citation3)
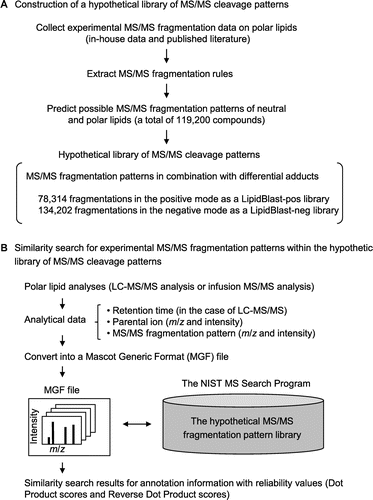
As an alternative to conventional lipid identification pipelines, we employed an in silico approach by means of the LipidBlast software package (http://fiehnlab.ucdavis.edu/projects/LipidBlast).Citation4–Citation6) LipidBlast is a freely available computer-generated hypothetical MS/MS fragment library covering various lipid species, including neutral and polar lipids (Fig. ). The fragmentation patterns were predicted by ion fragmentation and rearrangement rules generated from experimental MS/MS fragmentation data from in-house analyses and the literature. The package of MS/MS fragmentation patterns includes 212,516 MS/MS spectra (78,314 in positive mode as a LipidBlast-pos library and 134,202 in negative mode as a LipidBlast-neg library) for 119,200 structures. Spectral data from LC–MS/MS analyses are exported following the Mascot Generic Format (MGF), and are used for the similarity searches within the hypothetical spectral library by the NIST Mass Spectral Search Program (version 2.0). Search results are shown with match scores (Dot Product and Reverse Dot Product values) representing the reliability of cosine correlation (excellent, >900; good, 800–900; fair, 700–800; poor, <600).Citation7) In the similarity calculation, experimentally observed ions absent from the library are excluded from the calculation of Reverse Dot Product values. Thus, the LipidBlast search does not compare lipid accumulation levels among biological samples, but extrapolates the presence of lipid species from the in silico database of MS/MS fragmentation patterns.
Fig. 1. Lipid annotation scheme by means of the LipidBlast program.
Note: (A) Preparation of a hypothetical fragmentation pattern library. The LipidBlast program package (http://fiehnlab.ucdavis.edu/projects/LipidBlast/) contains the NIST Mass Spectral Search Program and a hypothetical MS/MS cleavage pattern library. For the hypothetical library, MS/MS cleavage rules of polar lipids were predicted based on experimentally obtained MS/MS fragmentation data on polar lipids (in-house data) and MS/MS fragmentation data on polar lipids from the published literature. The cleavage rules were applied to neutral and polar lipids (119,200 compounds). The hypothetical library consists of 78,314 positive MS/MS spectra (lipidblast-pos library) and 134,202 negative MS/MS spectra (lipidblast-neg library) with various adduct ions. (B) Execution of the LipidBlast program. Experimental MS/MS fragmentation data are used in a similarity search of the hypothetical MS/MS library. Neutral and polar lipid analyses are done by LC–MS/MS or infusion MS/MS. Analytical data (retention time, m/z and ion intensity information for parental ions, m/z and ion intensity information for MS/MS fragments) were converted into MGF files. For the LipidBlast program, the hypothetical MS/MS spectral library (lipidblast-pos or lipidblast-neg library) is installed into the NIST Mass Search Program. A similarity search by the LipidBlast yielded top hit lists of annotation information (lipid-head-group information, lipid carbon chain length information, lipid unsaturated bond number information, and detailed lipid structural information) with match scores for result reliability (Dot Product and Reverse Dot Product).
Here we present a rapid profiling of polar lipid accumulation in Euglena gracilis Z strain under anaerobic growth conditions by LipidBlast. Euglena, a unicellular eukaryotic flagellate belonging to the phylum Euglenophyta,Citation8) can grow under autotrophic, heterotrophic, or mixotrophic conditions. Most Euglena species are photoautotrophic owing to the presence of functional chloroplasts, which originate in the endosymbiosis of eukaryotic green algae.Citation9) The chloroplasts in Euglena contain pyrenoids that function as machinery specializing in efficient CO2 fixation and the synthesis of paramylon, a β-1,3-glucan that performs energy storage functions under adverse conditions.Citation10–Citation12) Hypoxia triggers wax ester fermentation, a quick metabolic shift favoring the syntheses of wax esters, odd-numbered fatty acids, and fatty alcohols,Citation13,Citation14) but possible changes in membrane lipid composition remain to be clarified during wax ester fermentation.
The E. gracilis Z strain was maintained in a modified Koren–Hutner medium (pH 5.0)Citation15) at 28 °C under continuous light on a rotary shaker (125 rpm). Wax ester fermentation was induced by halting culture agitation.Citation13) Cells were collected at 0 and 24 h after the initiation of anaerobiosis and were immediately immersed in liquid nitrogen, followed by lyophilization. The total lipid fraction was extracted by a modification of the Folch method with a chloroform/methanol (2:1) mixture.Citation16) The extracts were dissolved in methanol containing 5 μM reserpine as internal standard. MS/MS measurements (positive ion mode) were done by liquid chromatography linear ion trap time-of-flight mass spectrometry (LC-IT/TOFMS; Nano Frontier LD, Hitachi HighTechnologies, Tokyo). Samples were separated on a YMC-Pack Diol-120-NP column (150 × 2.0 mm, 5 μm particle; YMC, Kyoto, Japan) by a gradient elution method with solvent A (0.1% formic acid in water) and solvent B (0.1% formic acid in acetonitrile) at a flow rate of 0.2 mL/min and a column temperature at 40 °C. Hydrophilic interaction chromatography was done by the following elution program: 0 min, 3% A; 20 min, 35% A; 20.1 min, 50% A; 25 min, 50% A; 25.1 min, 3% A; 37 min, 3% A. Electrospray ionization (ESI) with positive mode was set to acquire a full MS scan between m/z 100 and 1,500, followed by MS/MS scans between m/z 400 and 1,500 of the top two ions detected in the full MS scan.
The profiles of the total ion chromatogram (TIC) exhibited slight differences between samples before (0 h) and after (24 h) anaerobic culture of the Z strain, suggesting that anoxia influenced polar metabolite compositions within 24 h (data not shown). Nile Red staining (Fig. (A)) was used to indicate anoxia-induced neutral lipid biosynthesis. In order to achieve polar lipid annotation before and after anaerobic culture, the MS/MS fragmentation data were collected from triplicate LC–MS/MS experiments (from both the 0 and the 24 h samples) and used in the similarity search of MS/MS fragments by LipidBlast. Spectral data from the LC–MS/MS analyses were exported as MGF and used in a similarity search of the hypothetical spectral library by means of the NIST Mass Spectral Search Program (version 2.0). A MS/MS similarity search was done with the following settings: precursor mass accuracy setting, off; product ion search accuracy, 0.4 Da. The reliability of the similarity search results is presented as match scores (Dot Product and Reverse Dot Product values) for each parent ion with individual MS/MS fragmentation patterns. Examples of the similarity search results are shown in Fig. .
Fig. 2. Comparison of annotated lipid species of the E. gracilis Z strain before and after anaerobic culture.
Note: Nile Red staining images of E. gracilis Z strain with (a) and without (b) anoxia treatment (A). Cells were sampled at 0 h and 24 h after the initiation of anaerobiosis and stained with Nile Red solution.Citation18) Light microscopic images (left panel) and fluorescent microscopic images (right panel) are shown. Fluorescent images were visualized with an IX71 inverted microscope with a fluorescent imaging unit (Olympus, Tokyo). These Euglena cells were used in LC-IT/TOFMS analyses and LipidBlast profiling. Lipid classes of annotated lipid species (B), carbon chain lengths of annotated lipid species (C), and unsaturated bond numbers of annotated lipid species (D). Vertical bars compare the numbers of lipid annotations deduced from the experimental MS/MS fragmentation patterns on the basis of triplicate analyses. The lipid annotations were classified into lipid classes (shown below). In individual samples, the number of annotations belonging to each lipid class is shown in the percentage of the total annotation numbers (73, 76, and 73 annotations from the 0-h samples, and 94, 88, and 81 annotations from the 24-h samples). The error bars represent the standard deviations of triplicate experiments (independent sample extraction, MS analyses, and LipidBlast profiling). Asterisks indicate significant differences as compared to 0-h anoxia samples (*p < 0.05, **p < 0.01; Student’s t-test). White and gray bars indicate 0-h and 24-h anoxia treatments, respectively. The lipid classes are as follows: CerP, ceramide-1-phosphates; DG, diacylglycerols; lysoPC, lysoglycerophosphocholines; lysoPE, lysoglycerophosphoethanolamines; MG, monoacylglycerols; MGDG, monogalactosyldiacylglycerols; PA, glycerophosphates; PC, glycerophosphocholines; PE, glycerophosphoethanolamines; plasmenyl-PC, plasmenyl-glycerophosphocholines; plasmenyl-PE, plasmenyl-glycerophosphoethanolamines; PS, glycerophosphoserines; SM, sphingomyelins; and TG, triacylglycerols. NA not assigned.
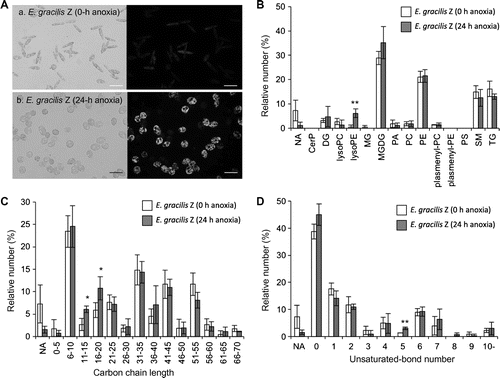
Fig. 3. Spectral similarity search results by the LipidBlast program (the library for positive mode analyses).
Note: Experimental MS/MS fragmentation data and retrieved hypothetical MS/MS fragmentation data are shown in the upper and lower panels, respectively. (A) A similarity search yielded the annotation lysoglycerophosphoethanolamine (lysoPE, 16:0/0:0) for experimental parent ion m/z 454. The annotation is shown with Dot Product and Reverse Dot Product values of 923 and 958, respectively. (B) A similarity search for the experimental parent ion m/z 831 yielded glycerophosphocholine (PC, 20:4/20:4) with the Dot Product and Reverse Dot Product values of 692 and 905, respectively. For this annotation, nine candidates with the same scores were generated due to the different positions of the double bonds.
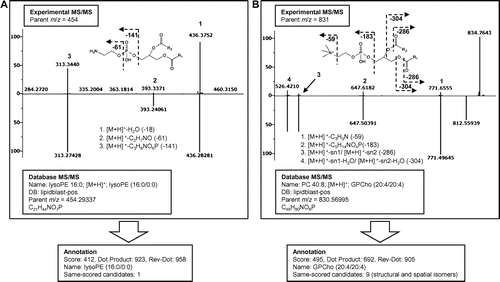
Fig. (B–D) show the LipidBlast results, comparing the annotation numbers of various lipid classes before (0 h) and after (24 h) anoxia. Compounds with low match score values (Dot Product value <600) were excluded from the comparison.
In summary, LipidBlast profiling for the triplicate experiments (from both the 0 and the 24 h samples) yielded 73, 76, and 73 lipid annotations and 94, 88, and 81 lipid annotations, respectively. The numbers of lipid species annotated as lysoglycerophosphoethanolamines (lysoPE) were significantly increased by anoxia, while those of the other lipid classes were unchanged (Fig. (B)). In general, lysoPE is produced by phospholipase A2 (PLA2), which catalyzes the hydrolysis of the sn-2 position of glycerophospholipids to yield free fatty acids and lysophospholipids.Citation17) Further studies are required to clarify the relationship between the carbon chain lengths of PE/lysoPE and the newly accumulated major wax ester components, which should be determined by GC–MS analyses. LipidBlast profiling also suggested that the lipid species of monogalactosyldiacylglycerols (MGDG) probably increased during the 24 h of anaerobic culture, while no statistically significant differences were confirmed (Fig. (B)). The carbon chain lengths and unsaturated bond numbers in each annotation were also compared (Fig. (C) and (D)). The numbers of lipid species with shorter carbon chain lengths (acyl chain lengths between 11 and 20) were increased markedly by anoxia (Fig. (C)). On the other hand, the numbers of unsaturated lipid species were unchanged, except for lipid species with five double bonds (Fig. (D)). These results suggest that anaerobiosis induces a dramatic metabolic shift not only in wax ester fermentation but also in phospholipid composition.
Global profiling of phospholipids is easily accomplished by means of robust lipid analytical platforms based on high-resolution MS systems and a series of standard reference compounds and in silico MS/MS libraries. We successfully demonstrated profiling of anoxia-induced phospholipid metabolism in E. gracilis by means of the LipidBlast package. Further information on carbon chain lengths and double bond positions can be confirmed by employing different approaches, including GC–MS analysis. The lipid annotation pipeline using such a virtual spectral fragmentation library offers a unique option for diverse research areas.
Funding
Funding was provided by the Strategic International Cooperative Program (SICP) – Joint Research Type Japanese (JST) – US (NSF) Joint Research “Metabolomics for a low carbon society” (METABOLOMICS).
T.K. and O.F. were also supported by the US National Science Foundation (MCB 1139644) and the US National Institutes of Health grants (P20 HL113452 and U24 DK097154).
Notes
Abbreviations: LC, liquid chromatography; MS, mass spectrometry; FT-ICR/MS, Fourier transform ion cyclotron resonance mass spectrometry; MGF, Mascot Generic Format; LC-IT/TOFMS, liquid chromatography linear ion trap time-of-flight mass spectrometry; ESI, electrospray ionization; TIC, total ion chromatogram; lysoPE, lysoglycerophosphoethanolamine; PLA2, phospholipase A2; MGDG, monogalactosyldiacylglycerol.
References
- Ivanova PT, Milne SB, Myers DS, Brown HA. Curr. Opin. Chem. Biol. 2009;13:526–531.
- German LB, Gillies LA, Smilowitz JT, Zivkovic AM, Watkins SM. Curr. Opin. Lipidol. 2007;18:66–71.
- Horai H, Arita M, Kanaya S, Nihei Y, Ikeda T, et al. J. Mass Spectrom. 2010;45:703–714.
- Kind T, Liu KH, Lee DY, DeFelice B, Meissen JK, Fiehn O. Nat. Methods. 2013;10:755–758.
- Kind T, Meissen JK, Yang D, Nocito F, Vaniya A, Cheng Y-S, VanderGheynst JS, Fiehn O. J. Chromatogr. A. 2012;1244:139–147.
- Meissen JK, Yuen BTK, Kind T, Riggs JW, Barupal DK, Knoepfler PS, Fiehn O. PLoS ONE. 2012;7:e46770.
- Stein SE. J. Am. Soc. Mass Spectrom. 1994;5:316–323.
- Guiry MD, Guiry GM. AlgaeBase [searched 2013 Aug 14]. Available from: http://www.algaebase.org.
- Gibbs SP. Can. J. Bot. 1978;56:2883–2889.
- Gibbs SP. J. Ultrastruc. Res. 1960;4:127–148.
- Osafune T, Yokota A, Sumida S, Hase E. Plant Physiol. 1990;92:802–808.
- Barsanti L, Vismara R, Passarelli V, Gualtieri P. J. Appl. Phycol. 2001;13:59–65.
- Inui H, Miyatake K, Nakano Y, Kitaoka S. FEBS Lett. 1982;150:89–93.
- Schneider T, Betz A. Planta. 1985;166:67–73.
- Koren LE, Hutner SH. J. Protozool. 1967; Supplement 14.
- Folch J, Lees M, Sloane Stanley GH. J. Biol. Chem. 1957;226:497–509.
- Kudo I, Murakami M. J. Biochem. 2002;131:285–292.
- Greenspan P, Mayer EP, Fowler SD. J. Cell Biol. 1985;100:965–973.
