Abstract
We have engineered a system that holds potential for use as a safety switch in genetically modified yeasts. Human apoptotic factor BAX (no homolog in yeast), under the control of the FBP1 (gluconeogenesis enzyme) promoter, was conditionally expressed to induce yeast cell apoptosis after glucose depletion. Such systems might prove useful for the safe use of genetically modified organisms.
In multicellular eukaryotes, apoptosis (a form of programmed cell death) is a process that eliminates unnecessary cells during embryonic development and in adult tissues.Citation1) Apoptotic cells digest themselves from the inside in a controlled manner, minimizing damage to neighboring cells.Citation1) Apoptotic cells are characterized by distinct morphological changes, Citation2) including phosphatidylserine exposure on the cell surface,Citation3) DNA cleavage,Citation4) nuclear fragmentation with chromatin condensation,Citation1), and fragmentation of subcellular organelles (the Golgi, endoplasmic reticulum, and mitochondrial networks).Citation5,6) Finally these cells break up into membrane-enclosed fragments called apoptotic bodies.Citation1) Among the proteins of the Bcl-2 family, some inhibit apoptosis, whereas others promote it.Citation7) One of the crucial pro-apoptotic proteins in the Bcl-2 family is the Bcl-2-associated X protein (Bax), which activates apoptosis by induction of cytochrome c release from the mitochondria.Citation7) Apoptosis also occurs in yeastCitation8) and bacteria. Saccharomyces cerevisiae lacks many of the key homologs of mammalian Bcl-2 family apoptotic regulators,Citation9) but overproduction of human Bax protein in yeast cells triggers typical apoptotic phenotypes (phosphatidylserine exposure, chromatin condensation, plasma membrane blebbing, and DNA fragmentation), leading to apoptosis of those cells.Citation10Citation−Citation12)
Genetically modified organisms (GMOs) are gaining importance in industry because they can be manipulated to produce required quantities of desired products under highly controlled conditions. The construction and propagation of GMOs is faster and less labor-intensive than traditional breeding methods. However, the use of GMOs is not as widespread as was expected, due to the environmental safety concerns. Since the release of GMOs into the general environment is prohibited, they must be inactivated or discarded with caution, severely limiting their application in industry. To overcome these limitations of GMO usage, we designed an apoptosis-inducing system that safely removes genetically modified yeast cells upon the completion of reactions. Since apoptosis causes DNA fragmentation, modified genes are inactivated in the reaction system, and the entire system can be discarded safely.
For glucose-dependent induction of apoptosis, the human BAX gene was placed under the control of the FBP1 promoter. FBP1 encodes fructose-1,6-bisphosphatase, a key enzyme that catalyzes in only one direction in the gluconeogenesis pathway.Citation13) Transcription of FBP1 is subject to catabolite repression in the presence of glucose as carbon source.Citation14) Upon glucose depletion, the transcription of FBP1 is considerably upregulated.Citation15) In this study, we constructed genetically modified yeast cells to undergo apoptosis on induction of human Bax expression controlled by the FBP1 promoter in response to glucose depletion.
To confirm whether the production of human Bax protein in yeast cells leads to apoptosis, the pYES3/CT (TRP1 marker) vector (InvitrogenTM, Life Technologies, Carlsbad, CA, USA), under the control of the galactose promoter, was used for conditional expression of the human BAX gene. DNA fragments encoding the EGFP gene were amplified by polymerase chain reaction (PCR) from pYEX-FETCitation16) by primers pYES3/CT-BamHI-XhoI-EGFP-F (5′-AGCTCGGATCCGAATTCCTCGAGGGTGGATCTGGTGGCATGGTGAGCAAGGGCGAGG-3′) and EGFP-SalI-pYES3CT-R (5′-CTAGAGTCGACTTACTTGTACAGCTCGTCCATGCC-3′). The PCR products were digested with BamHI and SalI and inserted into the BamHI-XholI sites of the pYES3/CT vector. The resulting plasmid was termed pYES3/CT-EGFP. Human BAX gene constructs, with and without stop codons, were amplified by PCR with human brain cDNA as template, with the following primer sets: (i) BAX-F-BglII (5′-ACTATAGATCTATGGACGGGTCCGGGG-3′) and BAX-R-XhoI (5′-GTTTCCTCGAGTCAGCCCATCTTCTTCCAGATG-3′) and (ii) BAX-F-BglII-2 (5′-ACTATAGATCTGCGGTGATGGACGGGTC-3′) and BAX-R-XhoI-outTGA (5′-GTTTCCTCGAGGCCCATCTTCTTCCAGATG-3′), respectively. The PCR products were inserted into the BamHI-XhoI sites of the pYES3/CT (no additional sequences) and pYES3/CT-EGFP (EGFP sequence fused at the C-terminus) vectors, and the resulting plasmids were named pYES3/CT-BAX and pYES3/CT-BAX-EGFP, respectively. These plasmids were introduced into S. cerevisiae strain W303-1A (MATa ade2-1 ura3-1 his3-11 trp1-1 leu2-3 leu2-112 can1-100).
Production of human Bax protein in yeast cells was confirmed by the emission of green fluorescence from EGFP (Fig. (A)). Cells were grown in SDC + AHLU medium containing 0.67% (w/v) yeast nitrogen base without amino acids, 2% glucose, 0.5% casamino acids, 0.002% l-adenine, 0.002% l-histidine, 0.003% l-leucine, and 0.002% uracil, and then inoculated into SGC + AHLU medium, in which galactose was used as a carbon source instead of glucose to induce expression by the galactose-driven promoter. After 6 h of induction, the cells were washed with phosphate-buffered saline (PBS, pH 7.4; 137 mm sodium chloride, 8.1 mm disodium hydrogen phosphate, 2.68 mm potassium chloride, and 1.47 mm potassium dihydrogen phosphate) and observed under an LSM 700 confocal microscope with ZEN software (Carl Zeiss, Jena, Germany).
Fig. 1. Induction of apoptosis by recombinant Bax production.
Note: (A) Production of human Bax driven by the galactose promoter in S. cerevisiae. BAX-EGFP denotes cells transformed with the pYES3/CT-BAX-EGFP plasmid, EGFP denotes cells with pYES3/CT-EGFP, and BAX denotes cells with pYES3/CT-BAX. Scale bar, 5 μm. (B) Viability of Bax-producing strains. CFUs of the Bax-producing strains (pYES3/CT-BAX, pYES3/CT-BAX-EGFP) and of control strains (pYES3/CT, pYES3/CT-EGFP) were counted after induction with galactose. Viability is represented as the percentage of CFUs obtained, as compared to the vector control, pYES3/CT. Error bars show standard deviations (n = 3). (C) Mitochondrial fragmentation observed by confocal microscopy. Mitochondria were stained by MitoTracker® Red CMXRos. Lower panel shows higher magnification of the images in the upper panel. Arrowheads indicate stained mitochondria. Scale bar, 5 μm.
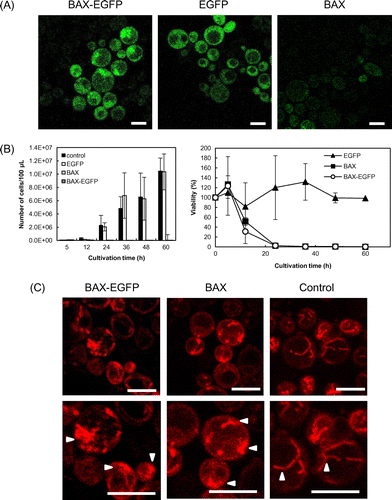
The viability of the cells producing human Bax was evaluated by counting the colony-forming units (CFUs). Cells were collected at appropriate time points after the induction of Bax in SGC + AHLU media, washed with PBS, serially diluted 10-fold from 10−1 to 10−5, and plated onto agar (2% w/v in media) plates. After 48 h of incubation, colonies were counted and percent viability was calculated by dividing the number of CFUs of the recombinant strains with the CFUs of the pYES3/CT (empty vector) strain. One hundred percent survival corresponded to the number of CFUs obtained with the vector control at each time point. As shown in Fig. (B), the viability of the pYES3/CT-BAX-EGFP and pYES3/CT-BAX strains decreased, whereas the viability of the cells with control vectors (pYES3/CT and pYES3/CT-EGFP) did not. These results indicate that recombinant human Bax was successfully produced by regulation of the GAL1 promoter, and triggered cell death in the yeast cells. To determine whether the cell death observed was apoptosis, the cells were examined for mitochondrial fragmentation. After 6 h of galactose induction, the cells were harvested and washed with PBS, and the mitochondria were stained with 100 nm MitoTracker® Red CMXRos (Molecular Probes®, Life Technologies). Red fluorescence was detected with the LSM 700 confocal microscope. As seen in Fig. (C), cells containing the control vector had intact mitochondrial network structures, whereas mitochondrial fragmentation was seen in cells producing Bax. These results confirm that the production of recombinant human Bax, with and without EGFP at the C-terminus, induced apoptosis in the yeast cells. However, previous studies have found that full-length unmodified human Bax cannot induce apoptosis,Citation17) and some tags at the C-terminus, e.g. a c-myc tag, have been thought to be necessary to induce Bax-induced apoptosis.Citation18) Our results, indicating that human Bax production without tags was sufficient to induce apoptosis, are inconsistent with a previous report of Priault M. et al.,Citation17) even though the experimental conditions were similar, the yeast strain (W303-1A) and the induction system with the GAL1 promoter (although using the pYES2/CT plasmid with URA3 marker, from Invitrogen) were the same. Differences in medium composition, especially galactose concentration, might have caused the difference in results. Priault et al. added 0.8% galactose to YNB medium (0.17% yeast nitrogen base, 0.5% ammonium sulfate, 0.1% potassium phosphate, 0.2% Drop-Mix, and 2% lactate, pH 5.5),Citation17) while we performed preculture in SDC + AHLU (glucose as carbon source), and then inoculation in SGC + AHLU containing 2% galactose. The higher concentrations of galactose in our culture medium might have contributed to the increase in Bax production.
Next, a system for apoptosis induction upon glucose depletion was constructed with the FBP1 promoter sequence and yeast integrating plasmid pRS404. The FBP1 promoter was cloned from the W303-1A genome with primer XhoI-FBP1-p-o-F (5′-GGAATCCCTCGAGTTCCGACCCTCTGTACTG-3′) and primer FBP1-p-BamHI-R (5′-ACTAGTGGATCCGTATGAGGGATGTTTCTTAT-3′), and inserted into the XhoI-BamHI sites of the pRS404 vector, yielding the pRS404-FBP1pro plasmid. Next, DNA sequences encoding EGFP-CYC1 terminator, Bax-CYC1 terminator, and Bax-EGFP-CYC1 terminator were inserted into pRS404. The EGFP-CYC1 terminator and BAX-EGFP-CYC1 terminator, BAX-CYC1 terminator, sequences were amplified from pYES3/CT-EGFP, pYES3/CT-BAX, and pYES3/CT-BAX-EGFP, respectively, and inserted into the BamHI-NotI sites of pRS404-FBP1pro with the following primers: pWI3-BglII-XhoI-EGFP-F (5′-ACTATAGATCTCCATGGACGCGTCTCGAGGGTGGATCTGGTGGCATGGTGAGCAAGGGCGAGG-3′), EGFP-NotI-o-p-R (5′-CGAGCTCGCGGCCGCTTACTTGTACAGCTCGTCCATGC-3′), BAX-F-BglII-2, and BEC-p-o-NotI-R (5′-CGAGCTCGCGGCCGCAGCTTGCAAATTAAAGCCTTCGA-3′). The resulting plasmids, termed pRS404-FBP1pro (empty vector), pRS404-FBP1pro-EGFP (encoding EGFP), pRS404-FBP1pro-BAX (encoding Bax), and pRS404-FBP1pro-Bax EGFP (encoding Bax-EGFP), were transformed into W303-1A.
Production of EGFP and Bax-EGFP, driven by the FBP1 promoter, was confirmed by the detection of fluorescence emitted by EGFP (Fig. (A)). Bax-EGFP exhibited a punctate distribution in the yeast cells. This pattern might be attributed to oligomerization of Bax-EGFP and its translocation to the mitochondrial outer membrane.Citation19) Bax-EGFP (Fig. (A)) showed massive distribution through the cells as compared to the cells producing EGFP. This may have been because Bax-EGFP (Fig. (A)), was strongly produced under the galactose-driven promoter.
Fig. 2. Induction of apoptosis by Bax produced under the control of the FBP1 promoter.
Note: (A) Production of human Bax protein in yeast cells. The FBP1 promoter and the BAX gene were integrated into the genome of W303-1A by means of the pRS404 vector. BAX-EGFP denotes cells harboring pRS404-FBP1pro-BAX-EGFP, EGFP denotes cells harboring pRS404-FBP1pro-EGFP, and BAX denotes cells harboring pRS404-FBP1pro-BAX. Scale bar, 5 μm. (B) Viability of Bax-producing cells in a low-glucose medium. Viability was calculated in the way described in Fig. (B), with the strain harboring pRS404-FBP1pro as control. Error bars show standard deviations (n = 4 up to 24 h).
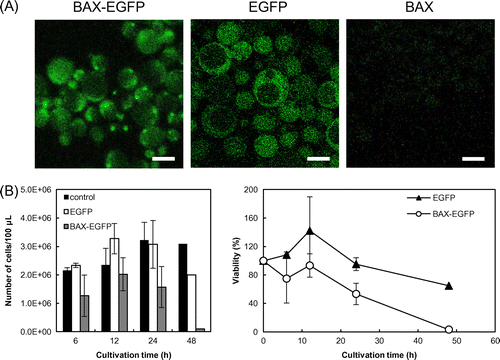
Next, we examined cell viability to determine whether the induction of BAX would trigger apoptosis. W303-1A yeast cells transformed with the pRS404-FBP1pro, the pRS404-FBP1pro-EGFP, or the pRS404-FBP1pro-BAX-EGFP plasmid were precultured in 10 mL of SDC + AHLU medium for 12 h, followed by inoculation into 10 mL of SD + AHLU (0.2% glucose) medium. Cells were collected after 6, 12, 24, and 48 h of incubation, and viability was evaluated by counting the CFUs. At 48 h, the viability of the cells expressing the pRS404-FBP1pro-BAX-EGFP plasmid decreased dramatically, to approximately 3%, as compared to that with the control plasmids pRS404-FBP1pro and pRS404-FBP1pro-EGFP (Fig. (B)). These results suggest that the FBP1 promoter responds to glucose depletion within 48 h by inducing Bax production, resulting in apoptosis of yeast cells.
Here, we report a system designed for inactivation of genetically modified yeast cells by means of a simple, non-destructive, glucose-depletion step, without the use of heat or filtration. Further studies should be done on aspects, such as optimizing the system to increase apoptotic rates at earlier time points, using multiple tandem recognition sites for increased transcription activation levels, Citation15) coexpression of other pro-apoptotic factors, and deletion of the endogenous Bax inhibitor, Bxi1p, in yeast.Citation20)
In conclusion, our genetically modified yeast system can be further extrapolated for other GMOs for applications in industrial bioconversion, without compromising the quality of the product or safety.
Acknowledgment
This work was supported by the commission for Development of Artificial Genes Synthesis Technology for Creating Innovative Biomaterial from the Ministry of Economy, Trade, and Industry (METI), Japan.
Notes
Abbreviations: BAX, BCL-2-associated X protein; CFU, colony-forming unit; GMO, genetically modified organism; PBS, phosphate-buffered saline; PCR, polymerase chain reaction.
References
- Kerr JF, Wyllie AH, Currie AR. Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br. J. Cancer. 1972;26:239–257.10.1038/bjc.1972.33
- Taylor RC, Cullen SP, Martin SJ. Apoptosis: controlled demolition at the cellular level. Nat. Rev. Mol. Cell Biol. 2008;9:231–241.10.1038/nrm2312
- Martin SJ, Reutelingsperger CP, McGahon AJ, Rader JA, van Schie RC, LaFace DM, Green DR. Early redistribution of plasma membrane phosphatidylserine is a general feature of apoptosis regardless of the initiating stimulus: inhibition by overexpression of Bcl-2 and Abl. J. Exp. Med. 1995;182:1545–1556.10.1084/jem.182.5.1545
- Wyllie AH, Kerr JF, Currie AR. Cell death: the significance of apoptosis. Int. Rev. Cytol. 1980;68:251–306.10.1016/S0074-7696(08)62312-8
- Frank S, Gaume B, Bergmann-Leitner ES, Leitner WW, Robert EG, Catez F, Smith CL, Youle RJ. The role of dynamin-related protein 1, a mediator of mitochondrial fission, in apoptosis. Dev. Cell. 2001;1:515–525.10.1016/S1534-5807(01)00055-7
- Lane JD, Lucocq J, Pryde J, Barr FA, Woodman PG, Allan VJ, Lowe M. Caspase-mediated cleavage of the stacking protein GRASP65 is required for Golgi fragmentation during apoptosis. J. Cell Biol. 2002;156:495–509.10.1083/jcb.200110007
- Youle RJ, Strasser A. The BCL-2 protein family: opposing activities that mediate cell death. Nat. Rev. Mol. Cell Biol. 2008;9:47–59.10.1038/nrm2308
- Madeo F, Frohlich E, Frohlich KU. A yeast mutant showing diagnostic markers of early and late apoptosis. J. Cell Biol. 1997;139:729–734.10.1083/jcb.139.3.729
- Fleury C, Pampin M, Tarze A, Mignotte B. Yeast as a model to study apoptosis? Biosci. Rep. 2002;22:59–79.10.1023/A:1016013123094
- Hanada M, Aime-Sempe C, Sato T, Reed JC. Structure-function analysis of Bcl-2 protein. Identification of conserved domains important for homodimerization with Bcl-2 and heterodimerization with Bax. J. Biol. Chem. 1995;270:11962–11969.
- Ligr M, Madeo F, Frohlich E, Hilt W, Frohlich KU, Wolf DH. Mammalian Bax triggers apoptotic changes in yeast. FEBS Lett. 1998;438:61–65.10.1016/S0014-5793(98)01227-7
- Priault M, Camougrand N, Chaudhuri B, Schaeffer J, Manon S. Comparison of the effects of bax-expression in yeast under fermentative and respiratory conditions: investigation of the role of adenine nucleotides carrier and cytochrome c. FEBS Lett. 1999;456:232–238.10.1016/S0014-5793(99)00957-6
- Entian KD, Vogel RF, Rose M, Hofmann L, Mecke D. Isolation and primary structure of the gene encoding fructose-1,6-bisphosphatase from Saccharomyces cerevisiae. FEBS Lett. 1988;236:195–200.10.1016/0014-5793(88)80313-2
- Zaragoza O, Vincent O, Gancedo JM. Regulatory elements in the FBP1 promoter respond differently to glucose-dependent signals in Saccharomyces cerevisiae. Biochem. J. 2001;359:193–201.10.1042/0264-6021:3590193
- Turcotte B, Liang XB, Robert F, Soontorngun N. Transcriptional regulation of nonfermentable carbon utilization in budding yeast. FEMS Yeast Res. 2010;10:2–13.10.1111/fyr.2010.10.issue-1
- Hara K, Ono T, Kuroda K, Ueda M. Membrane-displayed peptide ligand activates the pheromone response pathway in Saccharomyces cerevisiae. J. Biochem. 2012;151:551–557.10.1093/jb/mvs027
- Priault M, Cartron PF, Camougrand N, Antonsson B, Vallette FM, Manon S. Investigation of the role of the C-terminus of Bax and of tc-Bid on Bax interaction with yeast mitochondria. Cell Death Differ. 2003;10:1068–1077.10.1038/sj.cdd.4401270
- Manon S, Chaudhuri B, Guerin M. Release of cytochrome c and decrease of cytochrome c oxidase in Bax-expressing yeast cells, and prevention of these effects by coexpression of Bcl-xL. FEBS Lett. 1997;415:29–32.10.1016/S0014-5793(97)01087-9
- Renault TT, Grandier-Vazeille X, Arokium H, Velours G, Camougrand N, Priault M, Teijido O, Dejean LM, Manon S. The cytosolic domain of human Tom22 modulates human Bax mitochondrial translocation and conformation in yeast. FEBS Lett. 2012;586:116–121.10.1016/j.febslet.2011.12.003
- Cebulski J, Malouin J, Pinches N, Cascio V, Austriaco N. Yeast Bax inhibitor, Bxi1p, is an ER-localized protein that links the unfolded protein response and programmed cell death in Saccharomyces cerevisiae. PLoS ONE. 2011;6:e20882.10.1371/journal.pone.0020882
