Abstract
Transmembrane protein CD36 binds multiple ligands, including oxidized low-density lipoproteins (oxLDLs) and long-chain fatty acids (LCFAs). Our aim was to determine whether LCFAs compete with oxLDLs for binding to CD36. We addressed this issue by examining the inhibitory effect of LCFAs against the binding of Alexa-fluor-labeled oxLDLs (AFL-oxLDL) to a synthetic peptide representing the oxLDL-binding site on CD36 (3S-CD36150–168). All of the unsaturated LCFAs tested, inhibited the binding of AFL-oxLDL to 3S-CD36150–168, albeit to varying degrees. For instance, the concentrations required for 50% inhibition of binding for oleic, linoleic, and α-linolenic acids were 0.25, 0.97, and 1.2 mM, respectively. None of the saturated LCFAs tested (e.g. stearic acid) exhibited inhibitory effects. These results suggest that at least unsaturated LCFAs can compete with oxLDLs for binding to CD36. The study also provides information on the structural requirements of LCFAs for inhibition of oxLDLs–CD36 binding.
Graphical Abstract
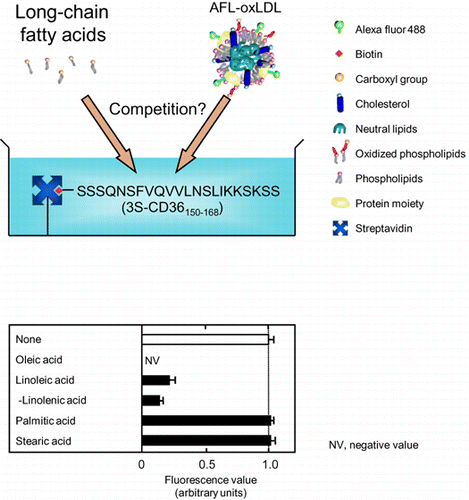
Unsaturated long-chain fatty acids but not saturated ones compete with Alexa-Fluor-labeled oxidized low-density lipoproteins for binding to a synthetic CD36 peptide.
CD36, is a transmembrane protein comprising of 472 amino acid residues that are expressed in a variety of cells, including monocytes/macrophages, and adipocytes.Citation1Citation−Citation3) It is known to mediate cellular uptake and the clearance of substances generated under oxidative stress, such as oxidized low-density lipoproteins (oxLDLs).Citation1−Citation3) oxLDL is known to serve as a high-affinity ligand of CD36, and hence its binding characteristics have been extensively explored. An important finding is that CD36-dependent recognition of oxLDL is mediated by oxidized glycerophospholipids (oxPLs), which are formed during the oxidation of LDL.Citation2,3) As a consequence of oxidation, an sn-2 acyl group of phospholipids on the surfaces of the LDL particles incorporates a terminal γ-hydroxyl(or oxo)-α,β-unsaturated carbonyl, which serves as a structural context for the binding of oxLDLs to CD36.Citation2,3)
A site within the extracellular domain of CD36 (amino acid positions 30 to 439) critical to oxLDL/oxPL binding has also been reported. For instance, of several synthetic peptides, one spanning amino acids 154 to 168 of human CD36 inhibited the binding of 125I-labeled oxLDLs to cultured mammalian cells that stably transfected with human CD36 cDNA.Citation4) We have found that (i) an oxLDL labeled with a fluorescence dye, Alexa Fluor® 488 (hereinafter AFL-oxLDL), bound specifically and saturably to streptavidin-coated plates that had been pretreated with a synthetic peptide corresponding to amino acids 149 to 168 of mouse CD36 with biotin at the N-terminus (CD36149–168, Fig. (A)), and (ii) the binding of AFL-oxLDL to the model CD36 was inhibited by oxPL species, such as 1-palmitoyl-2-(5-keto-6-octene-dioyl)phosphatidylcholine (KOdiA-PC), but not by a non-oxidized phospholipid species, 1-palmitoyl-2-arachidonoyl-sn-glycero-3-phosphocholine (PAPC).Citation5) These findings suggest that the region within the sequence of amino acids 149–168 serves as a binding site for oxLDLs/oxPLs.
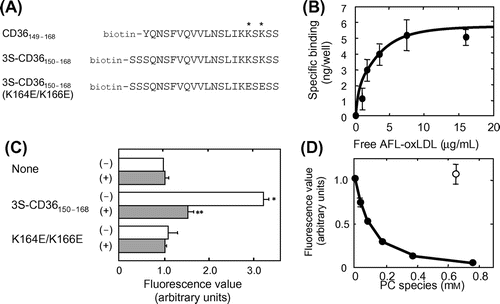
Fig. 1. Utility of 3S-CD36150–168 as a model CD36.
Note: (A) The amino acid sequences of CD36149–168, 3S-CD36150–168, and 3S-CD36150–168 (K164E/K166E) are shown in a single-letter code with biotin at the N-terminus. Lysine residues (Lys164 and Lys166), indispensable for interaction with oxLDL/oxPLs, are indicated by asterisks. (B) AFL-oxLDL binding to 3S-CD36150–168 but not to 3S-CD36150–168 (K164E/K166E). Wells of streptavidin-coated plates were pretreated with PBS-BSA alone (None), PBS-BSA containing 5 μM 3S-CD36150–168, or PBS-BSA containing 5 μM 3S-CD36150–168 (K164E/K166E) (K164E/K166E). They were then incubated with PBS-BSA containing 8 μg/mL of AFL-oxLDL in the absence (–) and in the presence (+) of 0.32 mg/mL of non-labeled oxLDL. The mean fluorescence value for the wells pretreated with PBS-BSA alone and then incubated with AFL-oxLDL alone was set at 1.0. Data are expressed as mean ± SD for at least two independent wells. They were analyzed by two-way analysis of variance and Bonferroni’s post hoc test. The p values obtained were 0.0002 for the effect of plate treatment, 0.0020 for the effect of non-labeled oxLDL, and 0.0009 for the plate treatment × non-labeled oxLDL interaction. *p < 0.001 vs. None (–). **p < 0.001 vs. 3S-CD36150–168 (–). p > 0.05 for None (–) vs. None (+), None (–) vs. K164E/K166E (–), and for K164E/K166E (–) and K164E/K166E (+). (C) Saturable pattern of specific binding of AFL-oxLDL to 3S-CD36150–168. Plots of the amounts of AFL-oxLDL bound to plates pretreated with 3S-CD36150–168 vs. the concentration of free AFL-oxLDL are shown. The curve was drawn with the aid of Prism®. Data are expressed as mean ± SD for three independent wells. (D) Inhibition of AFL-oxLDL binding to 3S-CD36150–168 by KOdiA-PC, but not by PAPC. Wells pretreated with PBS-BSA containing 5 μM 3S-CD36150–168 were incubated with PBS-BSA containing 8 μg/mL of AFL-oxLDL in the absence and the presence of KOdiA-PC at the concentrations indicated (solid circles) or PAPC at a concentration of 0.64 mM (hollow circle). Fluorescence values were obtained as described in the text. The mean fluorescence value obtained for the wells incubated with AFL-oxLDL alone was set at 1.0. Data are expressed as mean ± SD for at least two independent wells.
CD36 is also known to be involved in the cellular uptake, trafficking, and metabolism of long-chain fatty acids (LCFAs), and hence is often referred to as fatty acid translocase.Citation6Citation−Citation8) Binding between LCFAs and CD36 has also been shown by an assay using fatty acid binding resin Lipidex-1000.Citation7) To date, however, the details of LCFA binding to CD36, including the lipid-binding sites, remain largely unclear. This might be due to the difficulties in performing conventional binding assays using LCFAs as labeled ligands. It is not practical to add excess non-labeled LCFAs to assay mixtures to obtain values for specific binding.
It was recently found that (i) the rate of LCFA uptake was lower in Chinese hamster ovary cells expressing CD36 bearing mutation at Lys164 (located within the oxLDL-binding site) than in ones expressing wild-type CD36, and (ii) sulfo-N-succinimidyl oleate, which is believed to bind to CD36, inhibited oxLDL uptake by macrophages.Citation9) In addition, LCFAs and certain oxPLs, such as KOdiA-PC share common structural elements (a hydrocarbon chain and a carboxyl group).Citation2,3) Hence, we hypothesized that LCFAs can compete with oxLDLs/oxPLs for binding to CD36. The purpose of the present study was to address this hypothesis. To this end, we examined the inhibitory effect of LCFAs against the binding of AFL-oxLDL to a synthetic peptide representing the oxLDL-binding site on CD36. The data obtained suggest that at least certain unsaturated LCFAs can compete with oxLDLs/oxPLs for binding to CD36. Here we also provide information on the structural requirements of LCFAs for inhibition of oxLDL binding to CD36.
Materials and methods
Lipids
The purity of the lipids obtained from the manufacturers was > 98%. Arachidonic acid, linoleic acid, α-linolenic acid, γ-linolenic acid, methyl oleate, oleic acid, oleyl alcohol, palmitic acid, PAPC, and stearic acid were from Sigma (St. Louis, MO). Heptadecanoic acid was from Doosan Serdary Research Laboratories (Toronto, Canada). Eicosapentaenoic acid, ω-3 docosapentaenoic acid, ω-6 docosapentaenoic acid, and KOdiA-PC were from Cayman Chemical (Ann Arbor, MI). Docosahexaenoic, henicosanoic, nonadecanoic, 9E-octadecenoic acid, 6Z-octadecenoic acid, 11Z-octadecenoic acid, and tricosanoic acids were from Nu-Chek Prep (Elysian, MN).
Preparation of AFL-oxLDL
AFL-oxLDL was prepared with human oxLDL with a thiobarbituric acid-reactive substance value of about 30 nmoles of malondialdehyde/mg of protein (Biomedical Technologies, Stoughton, MA) and Alexa Fluor® 488 (Invitrogen, Carlsbad, CA), as described previously,Citation5) except that 0.5 mL of oxLDL solution, not having received dialysis, was added directly to a vial containing the reactive dye.
Pretreatment of streptavidin-coated plates with CD36 peptides and evaluation of the binding of AFL-oxLDL
Peptides conjugated with biotin at the N-terminus, 3S-CD36150–168 and 3S-CD36150–168 (K164E/K166E) (Fig. (A)), were synthesized by and purchased from Invitrogen. Each of the peptides was dissolved in dimethyl sulfoxide (DMSO) at a concentration of 1 mM, and was stored at –20°C until use.
The wells of a 96-well plate precoated with streptavidin (Thermo Fisher Scientific Japan, Tokyo) were prewashed three times with PBS containing 0.4% w/v BSA (hereinafter PBS-BSA). A 50-μL aliquot of PBS-BSA alone or of PBS-BSA containing 3S-CD36150–168 at a certain concentration (e.g. 5 μM) was added to each of the individual wells of the plate. Then the plate was incubated for 1 h at ambient temperature with gentle shaking. After it was washed thrice with PBS-BSA, each well received 50 μL of PBS-BSA containing AFL-oxLDL at a certain concentration (e.g. 8 μg/mL) in the presence and the absence of non-labeled oxLDL (e.g. 0.32 mg/mL). Then the plate was incubated in the dark for 1 h at ambient temperature with gentle shaking. After incubation, the wells were washed thrice with 0.2 mL of PBS and then exposed to 0.1 mL of PBS. The fluorescence in each well was measured with a plate reader (PowerScan, Dainippon Sumitomo Pharma, Osaka, Japan) at an excitation wavelength of 485 nm and an emission wavelength of 528 nm.
Determination of the dissociation constant (Kd) and the binding maximum (Bmax) of AFL-oxLDL binding to 3S-CD36150–168
The procedure for determining Kd and Bmax values was essentially the same as that described previously.Citation5) Wells pretreated with 5 μM 3S-CD36150–168 were exposed to 50 μL of PBS-BSA containing various concentrations of AFL-oxLDL in the absence and the presence of 0.32 mg/mL of non-labeled oxLDL, and the assay plate was incubated as described above. After incubation, the assay solutions were harvested and diluted 80-fold with PBS, and the fluorescence values (representing the concentrations of free ligand) were determined as described above. After harvesting of the assay solutions, the fluorescence of each well was measured as described above. The fluorescence value in each well was obtained by subtracting the mean background value of the wells that had been incubated with PBS-BSA alone. At each concentration of AFL-oxLDL, the value for specific binding (the amount of AFL-oxLDL bound to immobilized 3S-CD36150–168 in a well) was obtained by subtracting the mean fluorescence value in the presence of non-labeled oxLDL (non-specific binding) from the fluorescence value in the absence of non-labeled oxLDL (total binding). The values of Kd and Bmax were determined by fitting the progress curve to the theoretical curve by Equation (Equation1(1) ),Citation10 as follows, with the aid of commercially available software (Prism®, GraphPad, San Diego, CA):
(1)
where B is the amount of AFL-oxLDL bound to immobilized 3S-CD36150–168 in a well and [L] is the concentration of free AFL-oxLDL. The amounts of AFL-oxLDL bound to the peptide and the concentrations of free AFL-oxLDL were assessed by comparing them with the fluorescence values of the wells that received 0.1 mL of PBS containing AFL-oxLDL grains of known concentrations. The coefficient of determination (R2) was also obtained by means of Prism®.
oxLDL-binding competition assays
KOdiA-PC was supplied at a concentration of 1% (15 mM) in ethanol, and the other lipids tested in the present study were dissolved in ethanol at a concentration of 1%. The dilution series of lipid species were prepared as follows: A 10-μL aliquot of each lipid-containing solution was placed in a 1.5-mL microcentrifuge tube, to which was added 90 μL of a concentrated solution of PBS (1.1 × concentrate) containing 0.44% BSA, 44 μM butylated hydroxytoluene (BHT, Sigma), and 220 μM diethylenetriamine penta-acetic acid (DTPA, Dojindo, Kumamoto, Japan). The mixtures were diluted with PBS-BSA containing 10% ethanol, 40 μM BHT, and 200 μM DTPA to make a dilution series. Each of the dilution series was then diluted two-fold with PBS-BSA containing 16 μg/mL of AFL-oxLDL and then used in the experiments. Wells pretreated with 5 μM 3S-CD36150–168 were exposed to 0.1 mL of the dilution series of lipid species and the assay plate was incubated as described above. The fluorescence value for each well was measured as described above. The fluorescence values for the wells pretreated with 3S-CD36150–168 were obtained by subtracting the mean value of the peptide-untreated wells that had been incubated with 8 μg/mL of AFL-oxLDL. The concentration required for 50% inhibition of binding (the IC50 concentration) was determined by fitting the data to the theoretical curve by Equation (Equation2(2) ), as follows, with the aid of Prism®:
(2)
where Y is the amount of AFL-oxLDL bound to immobilized 3S-CD36150–168 in a well and [X] is the concentration of inhibitors (unsaturated LCFAs and KOdiA-PC). Note that this model assumes that the data have been normalized, and thus forces the curve to run from 100% down to 0%. For instance, the value for Y is set at 100 in the absence of inhibitors.
In the analysis to examine the effects of methyl oleate and oleyl alcohol, peptide-untreated wells as well as 3S-CD36150–168-treated ones were incubated with PBS-BSA containing 8 μg/mL of AFL-oxLDL in the absence and the presence of the test lipids. The effects were evaluated by comparing the values obtained by subtracting the mean values for the peptide-untreated wells from the values for the peptide-treated wells.
Statistical analysis
Data for the binding of AFL-oxLDL to the peptide-untreated and peptide-treated wells in the presence and the absence of non-labled oxLDL were analyzed by two-way analysis of variance and post hoc Bonferroni’s test by means of the aid of Prism®. The data for inhibition by LCFAs and oleic-acid derivatives (at a concentration of 0.05%) were analyzed by one-way analysis of variance and post hoc Bonferroni’s test by means of Prism®. A p-value of less than 0.05 was considered to be statistically significant.
Results
Preparation of a synthetic CD36 peptide and evaluation of the detection of AFL-oxLDL binding
We have confirmed the validity of CD36149–168 (Fig. (A)) in detecting AFL-oxLDL binding.Citation5) However, the use of CD36149–168 has a shortcoming in that the peptide initially dissolved in DMSO occasionally becomes insoluble when added to PBS-BSA. We have also found that a residue corresponding to Tyr149 of CD36 is not necessarily required for the detection of AFL-oxLDL binding.Citation11) In the present study, we prepared another CD36 peptide with potentially improved solubility, 3S-CD36150–168, which was comprised of amino acids 150 to 168 of mouse CD36 and had an additional three Ser residues on its N-terminal side (Fig. (A)). The three Ser residues were added in the expectation that they would contribute to improving the solubility of peptide. In our research, 3S-CD36150–168 has never become insoluble at any time when added to PBS-BSA.
We assessed to determine whether 3S-CD36150–168 can be used to detect AFL-oxLDL binding. The fluorescence values were significantly higher in the wells pretreated with 3S-CD36150–168 and then incubated with PBS-BSA containing AFL-oxLDL alone than in peptide-untreated control wells incubated with the same solution (Fig. (B)). As for the wells pretreated with 3S-CD36150–168, the fluorescence values were significantly lower in those incubated in the presence of non-labeled oxLDL than in those incubated in the absence of non-labeled oxLDL (Fig. (B)). These results indicate the specific binding interaction between 3S-CD36150–168 and AFL-oxLDL. Lys164 and Lys166 on CD36 have been found to be indispensable for binding interaction with oxLDL (Fig. (A)).Citation4) We have found that a CD36149–168 variant in which the residues corresponding to Lys164 and Lys166 were changed to Glu which exhibited poor or no ability to bind AFL-oxLDL.Citation5) The respective variant of 3S-CD36150–168 (designated 3S-CD36150–168 (K164E/K166E)) (Fig. (A)) produced similar results (Fig. (B)).
The specific binding of AFL-oxLDL to the plates pretreated with 5 μM 3S-CD36150–168 exhibited a dose-dependent saturation pattern (Fig. (C)). By fitting the progress curve of the amount of AFL-oxLDL bound to a well of the plate vs. the concentration of free AFL-oxLDL in the well to the theoretical curve by Equation (Equation1(1) ), the Kd and Bmax values were determined to be 2.3 μg/mL and 6.1 ng/well (R2 = 0.96), respectively. These values are similar to those obtained for plates pretreated with CD36149–168 at a concentration of 50 μM.Citation5)
An oxPL species, KOdiA-PC, inhibited binding of AFL-oxLDL to 3S-CD36150–168 in a concentration-dependent manner (Fig. (D)). The IC50 value was 0.10 mM (R2 = 0.97). PAPC exhibited no inhibition at a concentration of 0.64 mM. These results are similar to those obtained for CD36149–168.Citation5) We suggest that, like CD36149–168, 3S-CD36150–168 interacts with AFL-oxLDL by recognizing oxPL moieties.
The results presented in Fig. led us to conclude that 3S-CD36150–168 exhibits reliability and validity similar or even superior to those of CD36149–168 for detecting AFL-oxLDL binding.
Analysis of the inhibition of AFL-oxLDL binding to 3S-CD36150–168 by LCFAs
In this study, we used streptavidin-coated plates pretreated with 5 μM 3S-CD36150–168 to assess the inhibition of AFL-oxLDL binding by LCFAs. First, all of the tested LCFAs were added to the assay mixtures at a fixed concentration (0.05%) to assess comprehensively their inhibitory abilities. We included an antioxidant, BHT, and a metal chelator, DTPA (to remove trace levels of transition metal ions that might have catalyzed lipid oxidation during incubation) in the assay solutions.
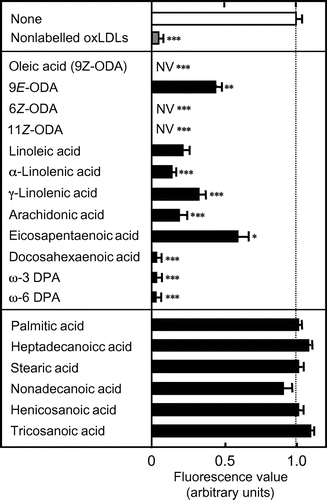
Compared to the fluorescence values in the wells incubated with the labeled ligand alone, the values were lower in the wells incubated with AFL-oxLDL in the presence of unsaturated LCFAs, oleic (18:1) (and the E isomer 9E-octadecenoic acid and the Z isomers 6Z- and 11Z-octadecenoic acids), linoleic (18:2), α-linolenic (18:3n-3), γ-linolenic (18:3n-6), arachidonic (20:4n-6), eicosapentaenoic (20:5n-3), docosahexaenoic (22:6), ω-3 docosapentaenoic (22:5n-3), and ω-6 docosapentaenoic (22:5n-6) acids (Fig. ). The fluorescence values were similar between wells incubated with AFL-oxLDL alone and those incubated with the labeled ligand in the presence of saturated LCFAs, palmitic (16:0), heptadecanoic (17:0), stearic (18:0), nonadecanoic (19:0), henicosanoic (21:0), and tricosanoic (23:0) acids (Fig. ). Statistical analysis revealed that the values for the wells incubated with unsaturated LCFAs were significantly different from those for the wells incubated with AFL-oxLDL alone, whereas those for the wells incubated with saturated LCFAs were not. Based on these results, we concluded that unsaturated LCFAs, but not saturated ones, inhibit the binding of AFL-oxLDL to 3S-CD36150–168. The fluorescence values were even lower in the wells incubated with AFL-oxLDL in the presence of oleic, 6Z-octadecenoic, and 11Z-octadecenoic acids than in the peptide-untreated wells incubated with the labeled ligand alone (i.e. the LCFAs produced negative values) (Fig. ). We interpret this to mean that these LCFAs at a concentration of 0.05% prevented even non-specific adsorption of AFL-oxLDL onto streptavidin and/or the assay plate wells.
Fig. 2. Comprehensive Analysis of the Inhibition of AFL-oxLDL Binding to 3S-CD36150–168 by LCFAs.
Note: Wells pretreated with 5 μM 3S-CD36150–168 were incubated with PBS-BSA containing 8 μg/mL of AFL-oxLDL in the absence and the presence of LCFAs at a concentration of 0.05%. Fluorescence values were obtained as described in the text. The mean fluorescence value for the wells pretreated with 3S-CD36150–168 and then treated with AFL-oxLDL alone was set at 1.0. Data are expressed as mean ± SD for three independent wells. Data were analyzed by one-way analysis of variance and Bonferroni’s post hoc test. *p < 0.05, **p < 0.01, and ***p < 0.001 vs. None. p > 0.05 for None vs. saturated LCFAs tested. DPA, docosapentaenoic acid; NV, negative value; ODA, octadecenoic acid.
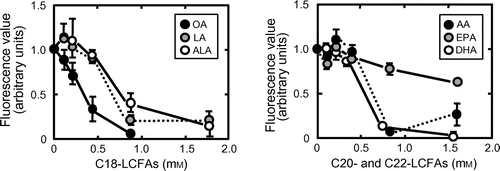
To assess more accurately the inhibitory abilities of unsaturated LCFAs, we investigated the concentration dependency of inhibition by them. As representatives, the data for oleic, linoleic, α-linolenic, arachidonic, docosahexaenoic, and eicosapentaenoic acids are shown (Fig. ). IC50 values were determined by fitting the data for the amount of AFL-oxLDL bound to a well of the plate vs. the logarithm of the concentration of LCFAs in the well to the theoretical curve by Equation (Equation2(2) ): oleic acid, 0.25 mM (R2 = 0.73); 9E-octadecenoic acid, 1.3 mM (R2 = 0.88); 6Z-octadecenoic acid, 0.48 mM (R2 = 0.71); 11Z-octadecenoic acid, 0.26 mM (R2 = 0.91); linoleic acid, 0.97 mM (R2 = 0.72); α-linolenic acid, 1.2 mM (R2 = 0.64); arachidonic acid, 0.70 mM (R2 = 0.63); eicosapentaenoic acid, 2.1 mM (R2 = 0.73); docosahexaenoic acid, 0.45 mM (R2 = 0.74); ω-3 docosapentaenoic acid, 0.35 mM (R2 = 0.80); and ω-6 docosapentaenoic acid, 0.30 mM (R2 = 0.77). Due to an unusual pattern of inhibition, (Supplemental Fig. S1; see Biosci. Biotechnol. Biochem. http://dx.doi.org/10.1080/09168451.2014.882750), the data for γ-linolenic acid could not be fitted to the modeled curve (R2 = 0.015). It remains unclear why γ-linolenic acid produced an unusual concentration-dependent pattern of inhibition. One possible explanation is that the LCFA at concentrations of about 0.9 mM adopts a conformation that is incompatible with binding to 3S-CD36150–168 in the assay mixture. Palmitic and stearic acids in concentration ranges of 0.13 to 2 and 0.11 to 1.8 mM, respectively, exhibited no inhibition.
Fig. 3. Inhibition of AFL-oxLDL binding to 3S-CD36150–168 in the presence of various concentrations of unsaturated LCFAs.
Note: Wells pretreated with 5 μM 3S-CD36150–168 were incubated with PBS-BSA containing 8 μg/mL of AFL-oxLDL in the absence and the presence of LCFAs at the concentrations indicated. Fluorescence values were obtained as described in the text. The mean fluorescence value for the wells pretreated with 3S-CD36150–168 and then treated with AFL-oxLDL alone was set at 1.0. Data are expressed as mean ± SD for three independent wells. AA, arachidonic acid; ALA, α-linolenic acid; DHA, docosahexaenoic acid; EPA, eicosapentaenoic acid; LA, linoleic acid; OA, oleic acid.
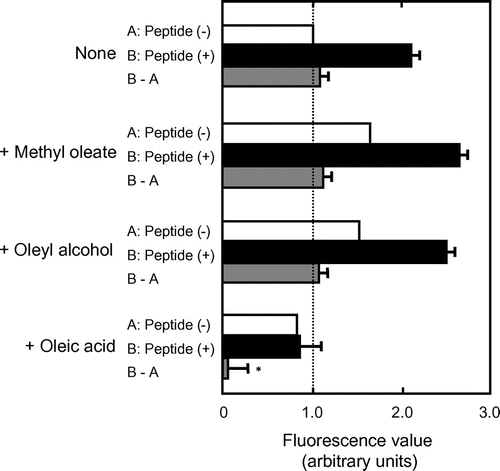
Analysis of inhibition of AFL-oxLDL binding to 3S-CD36150–168 by methyl oleate and oleyl alcohol
The observation that KOdiA-PC with the carboxyl end of the sn-2 chain inhibited AFL-oxLDL binding to 3S-CD36150–168, whereas PAPC, which lacks a carboxyl group, did not lead us to hypothesize that the carboxyl group in unsaturated LCFAs is important for inhibition. To test this hypothesis, we analyzed to determine whether methyl oleate and oleyl alcohol would inhibit binding.
The fluorescence values were higher in the peptide-untreated wells incubated with AFL-oxLDL in the presence of the oleic-acid derivatives than in those incubated with the labeled ligand alone (Fig. , white bars). We interpret this to mean that the oleic-acid derivatives increased non-specific adsorption of AFL-oxLDL onto streptavidin and/or the assay plate wells. A similar tendency was observed for the peptide-treated wells (Fig. , black bars). The values obtained by subtracting the mean values for the peptide-untreated wells from the values for peptide-treated wells were similar irrespective of the presence or absence of the derivatives (Fig. , gray bars). There were no statistically significant differences between the values from the wells incubated in the presence of oleic-acid derivatives and that from the well incubated with AFL-oxLDL alone. Based on these results, we conclude that the fatty acid derivatives had no ability to inhibit AFL-oxLDL binding to 3S-CD36150–168. The fluorescence values were slightly lower in the peptide-untreated wells incubated with AFL-oxLDL in the presence of oleic acid than in those incubated with the labeled ligand alone (Fig. ). These results support our interpretation that the fatty acid prevented even non-specific adsorption of AFL-oxLDL.
Fig. 4. Analysis of the Inhibition of AFL-oxLDL Binding to 3S-CD36150–168 by Methyl Oleate and Oleyl Alcohol.
Note: Wells pretreated PBS-BSA alone (A: Peptide (–), white bars) and those pretreated with PBS-BSA containing 5 μM 3S-CD36150–168 (B: Peptide (+), black bars) were incubated with PBS-BSA containing 8 μg/mL of AFL-oxLDL in the absence and the presence of methyl oleate, oleyl alcohol, or oleic acid at a concentration of 0.05%. The values obtained by subtracting the mean values for the peptide-untreated wells from the values for the peptide-treated wells (B–A) are indicated by gray bars. The mean fluorescence value for the peptide-untreated wells incubated with AFL-oxLDL alone was set at 1.0. Data are expressed as mean (white bars) and mean ± SD (black and gray bars) for three independent wells. The values for (B–A) were compared by one-way analysis of variance and Bonferroni’s post hoc test. *p < 0.001 vs. None (B–A). p > 0.05 for None (B–A) vs. methyl oleate and None (B–A) vs. oleyl alcohol (B–A).
Discussion
In this study, we used 3S-CD36150–168 as a model CD36. When using CD36149–168, it was necessary that streptavidin-coated plates are pretreated with the peptide at concentrations of 50 μM or more for successful detection of AFL-oxLDL binding.Citation5) When 3S-CD36150–168 was used, AFL-oxLDL binding was satisfactorily detected in assay plates pretreated with the peptide at concentrations of 5 μM or less (Y. Kozai, M. Takai, S. Tsuzuki, and T. Fushiki, unpublished results). In view of this improved solubility, 3S-CD36150–168 might be more effective at binding to streptavidin-coated plates than CD36149–168. The use of 3S-CD36150–168 provides a benefit in the low consumption of peptide in the assay.
Podrez et al. found that 0.1 mM oleic and linoleic acids caused no inhibition of 125I-labeled oxLDL binding to CD36-transfected human embryonic kidney 293 cells (and this is consistent with our finding that these LCFAs at concentrations around 0.1 mM produced no significant inhibition of AFL-oxLDL binding to 3S-CD36150–168; Fig. ) and suggested that LCFAs do not compete with oxLDLs/oxPLs for binding to CD36Citation2) but it remained to be determined whether LCFAs at concentrations higher than 0.1 mM exert inhibitory effects. Cultured cell-based assay systems might not be suitable to address this issue. In our research, oleic and linoleic acids, even at a concentration of 0.3 mM, appeared to cause membrane permeabilization of cultured cell lines (e.g. COS-1 monkey kidney cells), leading to CD36-independent entry of AFL-oxLDL into cells (that is, the production of a high-fluorescence background) (H, Inagaki, K. Kamei, M. Fujioka, S. Tsuzuki, and T. Fushiki, unpublished results). In the present study, we found that unsaturated LCFAs at a concentration of 0.5% (about 1.7 mM) exhibited inhibition of AFL-oxLDL binding to 3S-CD36150–168. To our knowledge, this is the first direct evidence suggesting that LCFAs (at least unsaturated ones) can inhibit the binding of oxLDLs/oxPLs to CD36. However, it remains uncertain whether this inhibition occurs under physiologically relevant conditions. In humans, serum concentrations of free fatty acids are known to exceed 1 mM in the postprandial state.Citation12) For instance, the serum concentration of free oleic acid is thought to exceed 0.3 mM in that state.Citation12) It is, thus, possible that at least certain unsaturated LCFAs (e.g. oleic acid) can inhibit oxLDL binding to CD36 under specific physiological conditions.
None of the saturated LCFAs tested inhibited AFL-oxLDL binding to 3S-CD36150–168. These results do not agree with previous findings that (i) palmitic and stearic acids (as well as oleic and arachidonic acids) bound to CD36 purified from rat adipocytes,Citation7) and (ii) the rate of palmitic acid uptake (as well as that of oleic-acid uptake) was lower in cells expressing CD36 bearing alanine substitution at Lys164 than in those expressing wild-type CD36.Citation9) A possible explanation of the lack of effect of saturated LCFAs in our assay is that the lipids had very low solubility in the buffer we used. Indeed, our assay solution turned very milky when saturated LCFAs initially dissolved in ethanol were added. We cannot exclude the possibility either that the oxLDL-binding site in the context of full-length CD36 allows access by saturated LCFAs, whereas 3S-CD36150–168 does not. Further modifications to the CD36 peptide, including the addition of more residues, may be required to assess the inhibition of AFL-oxLDL binding by saturated LCFAs.
The inhibitory effect of unsaturated LCFAs appears to depend on the spatial configuration of the hydrocarbon chain. This is indicated clearly by the observation that oleic acid inhibited AFL-oxLDL binding more effectively than did the E isomer, 9E-octadecenoic acid. The number of double bonds in the hydrocarbon chain (in the context of ones with the same number of carbon atoms in the chain) also appeared to affect inhibitory ability substantially. For instance, degrees of inhibition differed substantially between oleic, linoleic, and α-linolenic acids, and between arachidonic and eicosapentaenoic acids. We also found that the oleic-acid derivatives exhibited no inhibition of AFL-oxLDL binding to 3S-CD36150–168. We suggest (i) that the hydrocarbon chain in unsaturated LCFAs determines their ability to inhibit oxLDL binding to CD36 and (ii) that the carboxyl group in the lipids is necessary for their inhibitory effects.
In summary, we suggest that at least unsaturated LCFAs can compete with oxLDLs/oxPLs for binding to CD36, but it remains to be determined whether the LCFAs indeed bind to the oxLDL/oxPL-binding site on CD36, and if so, whether the lipids bind to the site in the same way as oxLDLs/oxPLs does. A study is underway to obtain crystals of LCFA– and oxPL–3S-CD36150–168 complexes, and this should provide evidence of direct interaction between LCFAs and CD36, and define the molecular bases for this interaction. Our present findings might provide a coherent interpretation of the results of crystallographic studies.
Supplemental material
The supplemental material for this paper is available at http://dx.doi.org/10.1080/09168451.2014.882750.
Supplemental Fig. 2 caption
Download MS Word (19 KB)Supplemental Fig. 1
Download PDF (53.4 KB)Supplemental Fig. 2
Download MS Power Point (152.5 KB)Acknowledgment
We thank Takahiko Amitsuka for valuable discussion. This study was supported by the Program for Promotion of Basic and Applied Researches for Innovations in Bio-oriented Industry.
Notes
Abbreviations: AFL, Alexa Fluor® 488-labeled; BHT, butylated hydroxytoluene; DTPA, diethylenetriamine penta-acetic acid; KOdiA-PC, 1-(palmitoyl)-2-(5-keto-6-octenedioyl)phosphatidylcholine; LCFAs, long-chain fatty acids; oxLDLs, oxidized low-density lipoproteins; oxPLs, oxidized glycerophospholipids; PAPC, 1-palmitoyl-2-arachidonyl-sn-glycero-3-phosphocholine.
References
- Collot-Teixeira S, Martin J, McDermott-Roe C, Poston R, McGregor JL. CD36 and macrophages in atherosclerosis. Cardiovasc. Res. 2007;75:468–477.10.1016/j.cardiores.2007.03.010
- Podrez EA, Poliakov E, Shen Z, Zhang R, Deng Y, Sun M, Finton PJ, Shan L, Gugiu B, Fox PL, Hoff HF, Salomon RG, Hazen SL. Identification of a novel family of oxidized phospholipids that serve as ligands for the macrophage scavenger receptor CD36. J. Biol. Chem. 2002;277:38503–38516.
- Hazen SL. Oxidized phospholipids as endogenous pattern recognition ligands in innate immunity. J. Biol. Chem. 2008;283:15527–15531.10.1074/jbc.R700054200
- Kar NS, Ashraf MZ, Valiyaveettil M, Podrez EA. Mapping and characterization of the binding site for specific oxidized phospholipids and oxidized low density lipoprotein of scavenger receptor CD36. J. Biol. Chem. 2008;283:8765–8771.10.1074/jbc.M709195200
- Tsuzuki S, Takai M, Matsuno Y, Kozai Y, Fujioka M, Kamei K, Inagaki H, Eguchi A, Matsumura S, Inoue K, Fushiki T. A synthetic peptide-based assay system for detecting binding between CD36 and an oxidized low-density lipoprotein. Biosci. Biotechnol. Biochem. 2013;77:132–137.10.1271/bbb.120625
- Su X, Abumrad NA. Cellular fatty acid uptake: a pathway under construction. Trends Endocrinol. Metab. 2009;20:72–77.10.1016/j.tem.2008.11.001
- Baillie AG, Coburn CT, Abumrad NA. Reversible binding of long-chain fatty acids to purified FAT, the adipose CD36 homolog. J. Membr. Biol. 1996;153:75–81.10.1007/s002329900111
- Fukuwatari T, Kawada T, Tsuruta M, Hiraoka T, Iwanaga T, Sugimoto E, Fushiki T. Expression of the putative membrane fatty acid transporter (FAT) in taste buds of the circumvallate papillae in rats. FEBS Lett. 1997;414:461–464.10.1016/S0014-5793(97)01055-7
- Kuda O, Pietka TA, Demianova Z, Kudova E, Cvacka J, Kopecky J, Abumrad NA. Sulfo-N-succinimidyl oleate (SSO) inhibits fatty acid uptake and signaling for intracellular calcium via binding CD36 lysine 164: SSO also inhibits oxidized low density lipoprotein uptake by macrophages. J. Biol. Chem. 2013;288:15547–15555.10.1074/jbc.M113.473298
- Pedersen JB, Lindup WE. Interpretation and analysis of receptor binding experiments which yield non-linear Scatchard plots and binding constants dependent upon receptor concentration. Biochem. Pharmacol. 1994;47:179–185.
- Takai M, Tsuzuki S, Matsuno Y, Kozai Y, Eguchi A, Matsumura S, Inoue K, Fushiki T. Assessment of key amino-acid residues of CD36 in specific binding interaction with an oxidized low-density lipoprotein. Biosci. Biotechnol. Biochem. 2013;77:1134–1137.10.1271/bbb.130072
- Chung BH, Hennig B, Simon Cho BH, Darnell BE. Effect of the fat composition of a single meal on the composition and cytotoxic potencies of lipolytically-releasable free fatty acids in postprandial plasma. Atherosclerosis. 1998;141:321–332.10.1016/S0021-9150(98)00168-3
