Abstract
A putative indole diterpene biosynthetic gene cluster composed of eight genes was identified in a genome database of Phomopsis amygdali, and from it, biosynthetic genes of fusicoccin A were cloned and characterized. The six genes showed significant similarities to pax genes, which are essential to paxilline biosynthesis in Penicillium paxilli. Recombinants of the three putative prenyltransferase genes in the cluster were overexpressed in Escherichia coli and characterized by means of in vitro experiments. AmyG is perhaps a GGDP synthase. AmyC and AmyD were confirmed to be prenyltransferases catalyzing the transfer of GGDP to IGP and a regular di-prenylation at positions 20 and 21 of paxilline, respectively. AmyD is the first know example of an enzyme with this function. The Km values for AmyD were calculated to be 7.6 ± 0.5 μM for paxilline and 17.9 ± 1.7 μM for DMAPP at a kcat of 0.12 ± 0.003/s.
Graphical Abstract
An enzyme catalyzing the regular di-prenylation at positions 20 and 21 of paxilline was identified in Phomopsis amygdali and its property was characterized with recombinant
The isoprenoid compounds found in nature to date, at more than 50,000 known examples, include industrially useful compounds such as flavors, antibiotics, and plant hormones, among others.Citation1–3) In some cases, isoprenoids are attached to other moieties, such as a polyketide,Citation4) indole/tryptophan,Citation5) or (iso)flavonoid,Citation6) or to phenazine moieties.Citation7,8) Prenylated indole alkaloids such as paspalitrem A,Citation9) aflatrem,Citation10) janthitrems,Citation11,12) shearinines,Citation13,14) penitrem A,Citation15) lolilline,Citation16) and lolitrems,Citation17) all of which are produced by filamentous fungi, are examples of this.Citation18) These compounds are derived from polyprenyl diphosphates and tryptophan or its derivatives, and they show diverse chemical structures and biological activities.Citation19–21)
Pioneering studies of biosynthetic genes and enzymes for indole-diterpenes were carried out by Scott et al. with Penicillium paxilli, a paxilline producer.Citation22) Recently, we characterized details of the functioning of each of the pax genes (paxG, paxC, paxM, paxB, paxP, and paxQ) by heterologous expression, by in vitro assay with recombinant enzymes, and by biotransformation.Citation23) Moreover, the prenylation of indoles/indole-3-glycerol phosphates by PaxC and stepwise regular di-prenylation at positions 21 and 22 of paxilline by PaxD was clearly confirmed by in vitro experiments with purified recombinant enzymes.Citation23,24)
We have also reported on the function of atmD, which is located in the aflatrem biosynthetic gene cluster in Aspergillus flavusCitation25) and encodes an enzyme with 32% amino acid identity to PaxD. It catalyzed the reverse mono-prenylation of paxilline at position 20 and position 21. Both AtmD and PaxD accepted paspaline, an intermediate of paxilline biosynthesis that has a structure similar to paxilline, but the regiospecificity and the regular/reverse specificity for prenylation were unexpectedly altered. Both AtmD and PaxD catalyzed the regular mono-prenylation of paspaline at position 21 and position 22.Citation26) These results suggest that fungal indole diterpene prenyltransferases have the potential to alter regio- and regular/reverse specificities for prenylation and are applicable in the synthesis of industrially useful compounds.
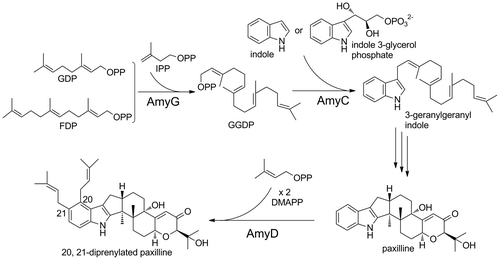
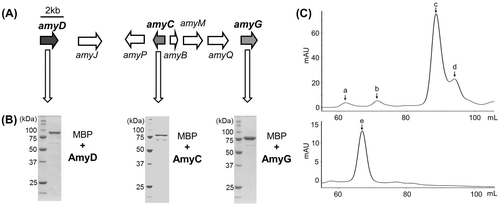
As part of our survey of a new fungal indole diterpene prenyltransferase, we identified a putative indole diterpene biosynthetic gene cluster composed of eight genes in the genome database of Phomopsis amygdali (Fig. (A)), a diterpene glucoside fusicoccin ACitation27) producer, for which we had identified the biosynthetic genes. The six genes (amyG, amyC, amyM, amyB, amyP, and amyQ) have significant similarities to the corresponding pax genes,Citation22) which are essential for paxilline biosynthesis. The remaining two genes are similar to prenyltransferases (amyD) and cytochrome P450s, suggesting that the cluster participates in the biosynthesis of a prenylated- and oxidized-paxilline such as lolilline and lolitrem. In this study, we investigated the substrate specificities of AmyD together with the PaxG and PaxC orthologs (AmyG and AmyC) with recombinant enzymes.
Fig. 1. The silent indole diterpene biosynthetic gene cluster identified in P. amygdali.
Note: Gene organization of amyD, amyC, and amyG (A), SDS-PAGE analysis of their recombinants (B), and gel filtration chromatography of recombinant AmyD (C). In (C), elution profiles of the standard proteins, aldolase (a, 158 kDa), albumin (b, 67 kDa), chymotrypsinogen A (c, 25 kDa), and ribonuclease A (d, 13.7 kDa); top and purified AmyD (bottom, e) are shown.
Materials and methods
General
Sequence analysis of the PCR fragments was performed by the dideoxy chain termination method with an automatic DNA sequencer (Li-Cor, Model 4000L, NE). Cell disruption was done with an Ultrasonic Disruptor (TOMY, UD-200, Tokyo). Analysis of the samples during protein purification was done by SDS-polyacrylamide gel electrophoresis (SDS-PAGE), and the proteins were visualized by Coomassie brilliant blue staining. The protein concentration was determined by the Bradford method, with bovine serum albumin as a standard. Plasmids from Escherichia coli were prepared with a Qiagen plasmid kit. All restriction enzymes, T4 ligase, and calf intestinal alkaline phosphatase were purchased from Toyobo (Osaka, Japan) and were used following the manufacturer’s instructions. Transformation of E. coli with plasmid DNA by electroporation was performed under standard conditions with a Gene Pulser electroporation system (Bio-Lad Japan, Tokyo).
cDNA preparation by heterologous expression in Aspergillus oryzae
The coding regions, including the intron sequences of the amyC, amyD, and amyG genes, were amplified by PCR with P. amygdali Niigata-2Citation28) genomic DNA and a primer set, as shown in Supplemental Table 1. The amplified DNA fragments were digested with the appropriate restriction enzymes and inserted into the corresponding sites of the expression vector, pTAex3,Citation29) to create pTAex3-amyG, pTAex3-amyC, and pTAex3-amyD. Then, each of the plasmids was introduced into A. oryzae.Citation23) Mycelia of the transformants were inoculated into a medium (3.5% Czapek-Dox, 2% starch, 1% polypeptone; final volume 100 mL) and cultivated at 30 °C for 3 d in a reciprocal shaker (125 strokes/min). cDNA preparation was performed by a method described previously,Citation30) with the primer set shown in Supplemental Table 1.
Overexpression and purification of prenyltransferases
After subcloning and sequence confirmation, the cDNA carrying the amyC, amyD, or amyG gene was inserted into the pMAL-c2X and into pMAL-c5X. In the resulting plasmids, pMAL-c2X-AmyC, pMAL-c5X-AmyD, and pMAL-c2X-AmyG, the recombinant protein was expressed as an N-terminal MBP-fused protein. The expression and purification conditions for the recombinant enzymes were essentially the same as those described in the manufacturer’s protocols. In brief, E. coli TB1 (New England Biolad, Japan, Tokyo), harboring the expression plasmid, was grown at 37 °C in Luria Broth medium with ampicillin (100 μg/mL) and D-glucose (0.2%). Expression of the recombinant protein was induced by adding 1 mM IPTG when the optical density at 600 nm reached about 0.8, and cultivation was continued for an additional 16 h at 18 °C. The recombinant protein was purified by amylose affinity column chromatography (New England Biolabs). After elution of the recombinant enzymes, purity was analyzed by SDS-PAGE on 10% gels. The protein concentration was determined by protein-dye standard assay (Bio-Rad) with bovine serum albumin as standard. For determination of the molecular mass and subunit structure, the purified enzyme was loaded onto a HiLoad 16/60 Superdex 75 pg gel-filtration column (Amersham Biosciences, Piscataway, NJ) and eluted with a buffer containing 50 mM Tris–HCl (pH 8.0) and 10 mM NaCl. The retention volume of AmyD was compared to those of the marker proteins. The markers applied to the column were aldolase (158 kDa), albumin (67 kDa), chymotrypsinogen A (25 kDa), and ribonuclease A (13.7 kDa) (GE Healthcare, Little Chalfont, UK).
In vitro assays of prenyltransferases
The assay mixture of AmyG in a final volume of 100 μL contained 0.5 mM of allylic substrates, 0.5 mM of IPP, 50 mM Tris–HCl (pH 8.0), 5 mM Mg2+ and a suitable amount of AmyG. The mixture was incubated at 30 °C overnight. After centrifugation at 15,000 rpm for 30 min, the precipitated products were solubilized with 100 μL of water and analyzed by high performance liquid chromatography (HPLC). The analytical conditions were as follows: Merck Mightisil RP-18GP Aqua column (250 mM × 4.6 mM) (Kanto Chemicals, Tokyo); mobile phase of acetonitrile in 10 mM potassium phosphate (pH 7.4) and 0.5% ion-pair reagents (0.5 mol/L, tetrabutylammonium dihydrogen phosphate solution; Wako, Osaka, Japan) (0 to 15 min, 40% acetonitrile; 15–30 min, 40–100%; 30–45 min, 100%); flow rate, 1.0 mL/min; detection, 205 nm. The assay methods for AmyC and AmyD were essentially as previously reported.Citation23,24,26)
The steady-state kinetic parameters of AmyD were determined by fitting to the Michaelis–Menten equation. The assay was linear with respect to the protein concentration at up to 1 μg for 30 min of incubation, and no substrate inhibition was observed for paxilline or DMAPP at up to 1.0 mM of a given substrate. The assays to determine the kinetic parameters of paxilline in a final volume of 100 μL contained 50 mM Tris–HCl (pH 8.0), 0.5 mM DMAPP, 0.2 μg of AmyD, and 1 μM to 0.5 mM paxilline. The mixture was incubated at 30 °C for 20 min. When the concentration of paxilline held steady at 0.25 mM, the concentration of DMAPP was varied from 5 μM to 0.5 mM.
Liquid chromatography–electrospray ionization mass spectrometry (LC/ESI-MS) analysis
Products formed in the in vitro assays were analyzed by LC/ESI-MS (Waters Acquity UPLC equipped with SQD2, Waters). The analytical conditions were as follows: Waters Acquity UPLC BEH C18 1.7 μm column (2.1 × 50 mM); column temperature, 40 °C; detection, positive mode; mobile phase, 0.1% formic acid: acetonitrile = 30:70 at 5 min, and a linear gradient to 50:50 for an additional 30 min; flow rate, 0.3 mL/min; cone voltage, 30 V.
Metal dependency of PAPT
Divalent metal ions (5 mM of Mg2+, Ca2+, Fe2+, Cu2+, Zn2+, Mn2+, Ni2+, and Co2+) or 5 mM EDTA was added to the standard reaction mixture.
Structural analysis of the reaction product
The reaction product formed from paxilline and DMAPP by AmyD was fractionated by a preparative HPLC. The 1H- and 13C-NMR spectra were recorded on a Bruker AMX-500 spectrometer: (Supplemental Table S2). NMR δH (CDCl3, 500 MHz) (Supplemental Fig. 1) 1.04 (s, 3H), 1.28 (s, 3H), 1.30 (s, 3H), 1.32 (s, 3H), 1.46 (m, 1H), 1.64 (m, 1H), 1.68 (s, 3H), 1.70 (s, 3H), 1.72 (s, 3H), 1.78 (m, 1H), 1.80 (s, 3H), 1.90 (m, 1H), 2.04 (m, 1H), 2.07 (m, 1H), 2.32 (m, 1H), 2.57 (m, 1H), 2.79 (m, 1H), 2.85 (m, 1H), 3.36 (d, J = 6.8 Hz, 2H), 3.61 (m, 2H), 3.73 (d, J = 2.0 Hz, 1H), 4.86 (brt, J = 9.7 Hz, 1H), 5.18 (m, 1H), 5.27 (m, 1H), 5.89 (d, J = 1.7 Hz 1H), 6.91 (d, J = 8.3 Hz 1H), 7.09 (d, J = 8.3 Hz 1H), 7.65 (s, 1H). NMR δC (CDCl3, 125 MHz) (Supplemental Fig. 2) 16.2, 17.9, 18.2, 19.7, 20.9, 24.2, 25.7, 25.8, 26.6, 28.0, 28.5, 29.1 (x2), 31.1, 34.4, 43.2, 49.5, 50.4, 72.5, 72.6, 77.6, 83.3, 109.4, 116.9, 119.6, 122.9, 124.4, 124.7, 125.3, 130.4 (x2), 130.6, 130.9, 138.4, 151.1, 168.2, 199.3. High resolution (HR)-ESIMS: calcd. for C37H50NO4 [M + H]+: 572.3734, observed: 572.3775.
Accession numbers
The nucleotide sequences reported here have been submitted to the DDBJ/GenBankTM/EBI Data Bank under Accession Nos. AB839408 (amyG), AB839409 (amyC), and AB839410 (amyD).
Results
Attempt to identify a compound governed by the cluster
Although the predicted amino acid sequences of AmyG and AmyC were similar to PaxG and PaxC, respectively, that of AmyD showed lower similarity to PaxD/AtmD (see below), and we were not sure whether the intrinsic substrate of AmyD is paxilline or another compound. In an attempt to identify the substrate of AmyD, we searched for an indole diterpene compound in the culture broth of P. amygdali, but no indole diterpene-related compounds were detected by LC-ESI-MS analysis under standard cultivation conditions. Trials to identify a compound related to indole diterpene by varying the culture medium and conditionsCitation31) and by adding histone deacetylase-inhibitors,Citation32,33) which are known to induce secondary metabolite genes, also failed. Next, an engineered strain from which the gene responsible for the first biosynthetic reaction of fusicoccin A (pafs gene) was deleted (Supplemental Fig. 3) to supply more IPP for indole diterpene biosynthesis was constructed because P. amygdali produced large amounts (>1 g/L) of fusicoccin-related compounds.Citation34) That strain, however, produced no specific compounds. Then, we focused on a transcriptional regulator, since the activation of transcriptional regulators sometimes results in high production of secondary metabolites in fungi,Citation35,36) but no candidate genes were located at the flanking regions of the cluster.
Preparation of cDNA by heterologous expression and overexpression of recombinant enzymes
Since we could not estimate the intrinsic substrate of AmyD, we attempted to prepare cDNAs by heterologous expression of it in A. oryzaeCitation37) and to examine the function of the recombinant enzyme with paxilline as substrate. The cDNAs of amyC and amyG were also prepared by the same method. Each amyC, amyD, and amyG gene was amplified by PCR and introduced into pTAex3, in which the inserted gene was expressed under the control of the promoter of the α-amylase gene of A. oryzae.Citation29) The transformants were cultivated, and then, cDNAs were prepared. The predicted gene products of AmyG, AmyC, and AmyD consisted of 339, 347, and 457 amino acids, respectively. The enzymes most closely homologous to AmyG and AmyC were PaxG (32% amino acid identity) and PaxC (40%), respectively, suggesting that the cluster participates in indole diterpene biosynthesis, but AmyD showed only low similarity to PaxD (13% amino acid identity), a prenyltransferase the gene locates in the paxilline biosynthetic gene cluster.
Each of the cDNAs was cloned into pMAL-c2X and pMAL-c5X vector for protein expression in E. coli. Maltose binding protein-fused recombinant enzymes were expressed successfully in soluble form under the conditions described above in “Materials and Methods”. The purified enzymes were subjected to SDS-PAGE to confirm their molecular sizes and purity (Fig. (B)). The recombinant enzyme of AmyD was then subjected to gel filtration to estimate its subunit structure. As shown in Fig. (C), a peak with a calculated molecular mass of 104 kDa was detected, suggesting that AmyD forms a monomer structure different from recombinant PaxD and AtmD, both of which showed homodimer structures, but this result might have been due to fusion of the large MBP at N-terminus, which is known to form a monomer structure.Citation38)
Functional analysis of AmyG and AmyC
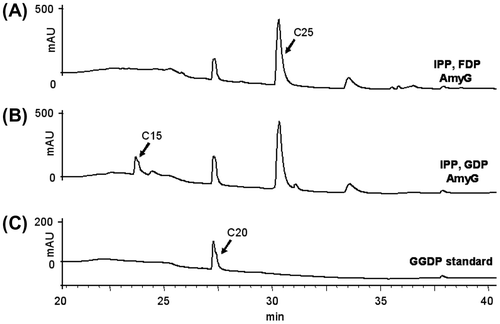
Catalytic activity was then examined with purified recombinant enzymes. Since AmyG was very similar to PaxG, the assay used for PaxG was employed. After recombinant AmyG was incubated with IPP and DMAPP, GDP, or FDP, the reaction product was analyzed by HPLC. When FDP and GDP were used, a peak that eluted at the same retention time as that of standard GGDP was detected together with another peak, which was farnesylgeranyl diphosphate (C25) in view of the retention times of FDP (C15) and GGDP, as shown in Fig. (A) and (B), but DMAPP was not utilized (data not shown). Considering that amyG clusters with many orthologs of the paxilline biosynthetic genes (Fig. (A) and Supplemental Table 3) and that the chain length of the product generated by the in vitro assay with GGDP synthase is known to vary depending on reaction conditions,Citation39) AmyG was perhaps a GGDP synthase.
Fig. 2. HPLC profiles of AmyG reaction products.
Note: (A) with IPP and FDP; (B) with IPP and GDP; C, GGDP standard.
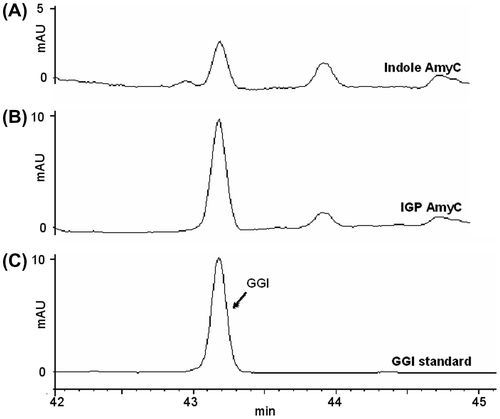
The enzymatic activity of AmyC was examined by the assay method used for PaxC.Citation23) Geranylgeranyl indole formed efficiently when AmyC was incubated with IGP and GGDP (Fig. (B)). Indole was also slightly accepted, in the same manner as PaxC (Fig. (A)). Although PaxC had been found to catalyze the transfer of FDP to IGP, yielding farnesyl indole, AmyC did not accept FDP (data not shown).
Functional analysis of AmyD
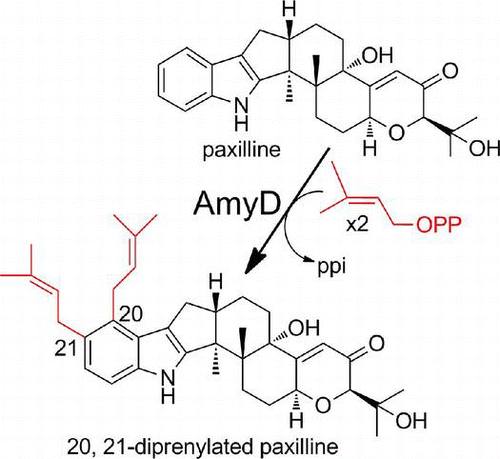
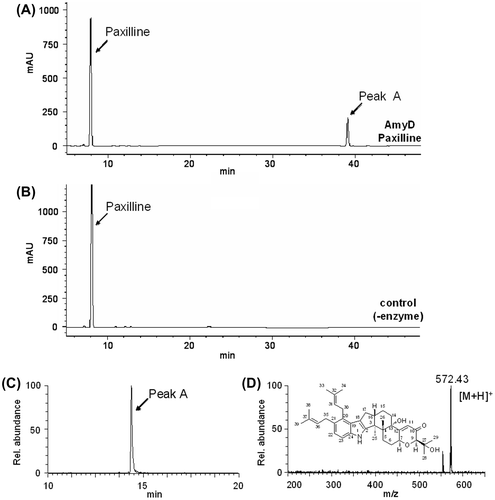
We characterized the recombinant AmyD using paxilline as substrate. The recombinant AmyD was incubated with paxilline and DMAPP and the reaction products were analyzed by HPLC, and a major product was specifically detected (Fig. (A)). Selected ion chromatograms obtained by LC/ESI-MS analysis showed a specific peak with a molecular mass corresponding to di-prenylated paxilline (Fig. (C) and (D)). HR (high resolution)-ESIMS of the product indicated the molecular formula C37H49NO4, confirming the production of di-prenylated paxilline. The 1H-NMR spectra showed the characteristic signals of regular prenyl moieties: two olefinic protons, 5.18 (m, 1H) and 5.27 ppm (m, 1H); two allylic methylene protons, 3.61 (m, 2H) and 3.36 (d, J = 6.8 Hz, 2H); and for the 4,5-di-substituted indole signals, aromatic protons, 6.91 (d, J = 8.3 Hz, 1H) and 7.09 ppm (d, J = 8.3 Hz, 1H)], indicating that the substitution of the prenyl moieties at the C-20 and C-21 positions on the indole moiety. Finally, extensive NMR data analysis, including hetero-nuclear single quantum coherence (HSQC) (Supplemental Fig. 4), hetero-nuclear multiple quantum coherence (HMBC) (Supplemental Fig. 5), correlation spectroscopy (COSY) (Supplemental Fig. 6), and nuclear overhauser effect spectroscopy (NOESY) (Supplemental Fig. 7) indicate that the structure was 20,21-di-prenylated paxilline (Supplemental Fig. 8). To the best of our knowledge, AmyD is the first enzyme known to catalyze the di-prenylation of paxilline at positions 20 and 21.
Fig. 4. HPLC and LC/ESI-MS analysis of the products formed in the reaction of paxilline and DMAPP catalyzed by AmyD.
Note: The reaction products formed with (A, C) and without (B) AmyD were analyzed by HPLC (A, B) and LC/ESI-MS (C, D). Selected ion chromatograms (C) and spectra of the peak (D) are shown.
Next, we tested paspaline, an intermediate of paxilline biosynthesis that has a structure similar to paxilline, as substrate, because both PaxD and AtmD have been found to accept this compound. The reaction products were analyzed by HPLC, and three small peaks were detected. By LC/ESI-MS analysis, one peak was perhaps a di-prenylated paspaline, and the other two were mono-prenylated paspalines (Supplemental Fig. 9). We could not determine the structure of these compounds due to low yield. However, the following observations suggested that the latter product might be 20-, 21-, or 22-mono-prenylated paspaline. The retention time of one of the mono-prenylated paspalines (Supplemental Fig. 9A, peak B) was the same as that of the 21- or 22-mono-prenylated paspaline, which has been found to form the same substrate by both AtmD and PaxD.Citation26) Both compounds were eluted at the same retention time by conventional reverse-phase column. The product formed by AmyD was also found to be a mixture of the 21- and 22-mono-prenylated paspaline by XBridgeTM Phenyl Column analysis (Supplemental Fig. 10). Since we did not access the standard compound of 20-mono-prenylated paspaline, we could not examine the retention time of this compound by the XBridgeTM Phenyl Column analysis. Considering that AmyD catalyzed prenylation at the 20 and 21 positions rather than the 21 and 22 positions, prenylation at the 20 position was also highly probable.
Biochemical characterization of AmyD
The biochemical properties of AmyD were investigated with paxilline and DMAPP as substrates. The optimal temperature and pH values were 55 °C and about 7.0 (Supplemental Figs. 11 and 12). Since some prokaryotic prenyltransferases require divalent cations for their activity, the effects and dependence of divalent metal ions on AmyD activity were tested (Supplemental Fig. 13). The addition of Mg2+ and Ca2+ enhanced enzymatic activity in contrast to EDTA, which decreased the activity to less than 50% of control. Activity was lost when Cu2+ and Zn2+ were added.
The kinetic parameters of AmyD were also investigated (Supplemental Fig. 14). The enzyme reaction followed Michaelis–Menten kinetics. By means of Hanes–Woolf plots, the Km values were determined to be 7.6 ± 0.5 μM for paxilline and 17.9 ± 1.7 μM for DMAPP. The kcat value was 0.12 ± 0.003/s.
Discussion
We identified a putative indole diterpene biosynthetic gene cluster composed of eight genes in the genome database of P. amygdali (Fig. (A)), from the strain we cloned the biosynthetic genes of fusicoccin A.Citation34) We attempted to identify a compound governed by the cluster, but all attempts failed. The following results, however, suggest that the cluster might be responsible for the biosynthesis of a lolilline/lolitrem type of compound. The six genes (amyG, amyC, amyM, amyB, amyP, and amyQ) have significant similarities to the corresponding pax genes,Citation22) which are essential for paxilline biosynthesis. Indeed, AmyG and AmyC were perhaps a GGDP synthase and a geranylgeranyl indole synthase, respectively, both of which are essential enzymes in the biosynthesis of indole diterpene-related compounds (Fig. ). Moreover, AmyD showed 58% amino acid identity to the product of ltmE, which has been identified in the lolitrems biosynthetic gene cluster in Epichloë festucae, in contrast its low similarity to AtmD and PaxD (17% and 13%). Scott et al. examined the function of the ltmE gene by constructing a gene-deletion mutant, introducing the mutant into its host plant, and performing structural analysis of a lolitrem-related compound produced by the endophyte. They suggested that LtmE catalyzes the di-prenylation of the A-ring of terpendole I, a paxilline-related compound, at positions, 20 and 21.Citation40) In the present study, AmyD was found to catalyze regular di-prenylation at the same positions of paxilline (Fig. ). Furthermore, three P450 monooxygenase genes are present in the amy gene cluster. In a search for homologous enzymes, two of them, AmyP and AmyQ, were found to show similarity to PaxP/LtmP (28%/34% identity) and PaxQ/LtmQ (22%/27%),Citation22,40) which catalyze successive reactions converting paspaline to paxilline. The other one showed high similarity to LtmJ (64% identity), a cytochrome P450 monooxygenase, which has been reported to be responsible for lolitremanes formation.Citation40)
In sum, it is suggested that the cluster identified in this study participates in the generation of a lolilline/lolitrem type of compound. To confirm this, the same strategy used for functional analysis of the paxilline biosynthetic gene cluster, the introduction of the pax genes into A. oryzae and an analysis of the compound produced by the transformant,Citation23) might be also useful.
Supplemental material
The supplemental material for this paper is available at http://dx.doi.org/10.1080/09168451.2014.882759.
Supplemental information - Tables S1-S3 and Figures S1-S14
Download MS Word (1.2 MB)Acknowledgments
This study was supported by Grants-in-Aid for Scientific Research (23108101 to T. Dairi and 22108002 to H. Oikawa) from the Japan Society for the Promotion of Science (JSPS).
Notes
Abbreviations: IPP, isopentenyl diphosphate; DMAPP, dimethylallyl diphosphate; GGDP, geranylgeranyl diphosphate; FDP, farnesyl diphosphate; GDP, geranyldiphosphate; IGP, indole-3-glycerol phosphate.
References
- Christianson DW. Science. 2007;316:60–61.10.1126/science.1141630
- Connolly JD, Hill RA. Dictionary of Terpenoids. New York (NY): Chapman and Hall; 1992.
- Dewick PM. Nat. Prod. Rep. 2002;19:181–222.10.1039/b002685i
- Lo HC, Entwistle R, Guo CJ, Ahuja M, Szewczyk E, Hung JH, Chiang YM, Oakley BR, Wang CC. J. Am. Chem. Soc. 2012;134:4709–4720.10.1021/ja209809t
- Steffan N, Grundmann A, Yin WB, Kremer A, Li SM. Curr. Med. Chem. 2009;16:218–231.10.2174/092986709787002772
- Tahara S, Ibrahim RK. Phytochemistry. 1995;38:1073–1094.10.1016/0031-9422(94)00788-U
- Izumikawa M, Khan ST, Takagi M, Shin-ya K. J. Nat. Prod. 2010;73:208–212.10.1021/np900747t
- Saleh O, Haagen Y, Seeger K, Heide L. Phytochemistry. 2009;70:1728–1738.10.1016/j.phytochem.2009.05.009
- Bills GF, Giacobbe RA, Lee SH, Peláez F, Tkacz JS. Mycol. Res. 1992;96:977–983.10.1016/S0953-7562(09)80601-1
- Gallagher RT, Clardy J, Wilson BJ. Tetrahedron Lett. 1980;21:239–242.10.1016/S0040-4039(00)71178-6
- de Jesus AE, Steyn PS, Van Heerden FR, Vleggaar R. J. Chem. Soc., Perkin Trans. 1 1984;1:697–701.10.1039/p19840000697
- Penn J, Swift R, Wigley LJ, Mantle PG, Bilton JN, Sheppard RN. Phytochemistry. 1993;32:1431–1434.10.1016/0031-9422(93)85153-I
- Belofsky GN, Gloer JB, Wicklow DT, Dowd PF. Tetrahedron. 1995;51:3959–3968.10.1016/0040-4020(95)00138-X
- Smetanina OF, Kalinovsky AI, Khudyakova YV, Pivkin MV, Dmitrenok PS, Fedorov SN, Ji H, Kwak JY, Kuznetsova TA. J. Nat. Prod. 2007;70:906–909.10.1021/np060396d
- de Jesus AE, Steyn PS, van Heerden FR, Vleggaar R, Wessels PL, Hull WE. J. Chem. Soc., Perkin Trans. 1. 1983;1:1847–1856.10.1039/p19830001847
- Munday-Finch SC, Wilkins AL, Miles CO, Tomoda H, Ōmura S. J. Agric. Food. Chem. 1997;45:199–204.10.1021/jf960396r
- Gallagher RT, Hawkes AD, Steyn PS, Vleggaar R. J. Chem. Soc. Chem. Commun. 1984;9:614–616.10.1039/c39840000614
- Li SM. Nat. Prod. Rep. 2009;27:57–78.10.1039/b909987p
- Dalziel JE, Finch SC, Dunlop J. Toxicol. Lett. 2005;155:421–426.10.1016/j.toxlet.2004.11.011
- González MC, Lull C, Moya P, Ayala I, Primo J, Primo Yúfera E. J. Agric. Food. Chem. 2003;51:2156–2160.10.1021/jf020983e
- Singh SB, Ondeyka JG, Jayasuriya H, Zink DL, Ha SN, Dahl-Roshak A, Greene J, Kim JA, Smith MM, Shoop W, Tkacz JS. J. Nat. Prod. 2004;67:1496–1506.10.1021/np0498455
- Young C, McMillan L, Telfer E, Scott B. Mol. Microbiol. 2001;39:754–764.10.1046/j.1365-2958.2001.02265.x
- Tagami K, Liu C, Minami A, Noike M, Isaka T, Fueki S, Shichijo Y, Toshima H, Gomi K, Dairi T, Oikawa H. J. Am. Chem. Soc. 2013;135:1260–1263.10.1021/ja3116636
- Liu C, Noike M, Minami A, Oikawa H, Dairi T. Appl. Microbiol. Biotechnol. 2014;98:199–206.
- Nicholson MJ, Koulman A, Monahan BJ, Pritchard BL, Payne GA, Scott B. Appl. Environ. Microbiol. 2009;75:7469–7481.10.1128/AEM.02146-08
- Liu C, Minami A, Noike M, Toshima H, Oikawa H, Dairi T. Appl. Environ. Microbiol. 2013;79:7298–7304.
- Ballio A, Brufani M, Casinovi CG, Cerrini S, Fedeli W, Pellicciari R, Santurbano B, Vaciago A. Experientia. 1968;24:631–635.10.1007/BF02153818
- Tajima N, Kume H, Kanematsu S, Kato N, Sassa T. J. Pestic. Sci. 2001;27:64–67.
- Fujii T, Yamaoka H, Gomi K, Kitamoto A, Kumagai C. Biosci. Biotechnol. Biochem. 1995;59:1869–1874.10.1271/bbb.59.1869
- Noike M, Liu C, Ono Y, Hamano Y, Toyomasu T, Sassa T, Kato N, Dairi T. Chembiochem. 2012;13:566–573.10.1002/cbic.v13.4
- Sanchez JF, Chiang YM, Szewczyk E, Davidson AD, Ahuja M, Elizabeth Oakley C, Woo Bok J, Keller N, Oakley BR, Wang CC. Mol. Biosyst. 2010;6:587–593.10.1039/b904541d
- Williams RB, Henrikson JC, Hoover AR, Lee AE, Cichewicz RH. Org. Biomol. Chem. 2008;6:1895–1897.10.1039/b804701d
- Henrikson JC, Hoover AR, Joyner PM, Cichewicz RH. Org. Biomol. Chem. 2009;7:435–438.10.1039/b819208a
- Noike M, Ono Y, Araki Y, Tanio R, Higuchi Y, Nitta H, Hamano Y, Toyomasu T, Sassa T, Kato N, Dairi T. PLoS One. 2012;7:e42090.10.1371/journal.pone.0042090
- Bok JW, Keller NP. Eukaryot. Cell. 2004;3:527–535.10.1128/EC.3.2.527-535.2004
- Yin W, Keller NP. J. Microbiol. 2011;49:329–339.
- Chiba R, Minami A, Gomi K, Oikawa H. Org. Lett. 2013;15:594–597.10.1021/ol303408a
- Spurlino JC, Lu GY, Quiocho FA. J. Biol. Chem. 1991;266:5202–5219.
- Matsuoka S, Sagami H, Kurisaki A, Ogura K. J. Biol. Chem. 1991;266:3464–3468.
- Saikia S, Takemoto D, Tapper BA, Lane GA, Fraser K, Scott B. FEBS Lett. 2012;586:2563–2569.10.1016/j.febslet.2012.06.035