Abstract
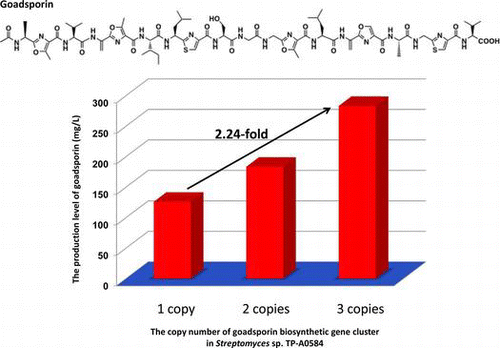
Improving the productivity of secondary metabolites is highly beneficial for the utilization of natural products. Here, we found that gene duplication of the goadsporin biosynthetic gene locus resulted in hyper-production. Goadsporin is a linear azole containing peptide that is biosynthesized via a ribosome-mediated pathway in Streptomyces sp. TP-A0584. Recombinant strains containing duplicated or triplicated goadsporin biosynthetic gene clusters produced 1.46- and 2.25-fold more goadsporin than the wild-type strain. In a surrogate host, Streptomyces lividans, chromosomal integration of one or two copies of the gene cluster led to 342.7 and 593.5 mg/L of goadsporin production. Expression of godI, a self-resistance gene, and of godR, a pathway-specific transcriptional regulator, under a constitutive promoter gave 0.79- and 2.12-fold higher goadsporin production than the wild-type strain. Our experiments indicated that a proportional relationship exists between goadsporin production per culture volume and the copy number of the biosynthetic gene cluster.
Graphical Abstract
Proportional relationship exists between Streptomyces antibiotic production and the copy number of the biosynthetic gene cluster.
Poor yields of desired bioactive natural products often limit drug discovery and development. Achieving high microbial production of complex natural products should offer advantages as to these limitations. Stable amplification of entire antibiotic biosynthetic gene clusters would be valuable for the improvement of antibiotic productivity. For example, it has been found that ZouA-mediated DNA amplification can be applied for enhancement of actinorhodin production in Streptomyces coelicolor A3(2).Citation1)
Our group has developed a series of integration vectors for actinomycetes enabling the heterologous expression of natural bacterial products.Citation2–4) In the present study, using these integration vectors, we tested how the duplication of a whole natural product biosynthetic gene cluster affects the total production of natural products.
Goadsporin is a linear azole containing peptide (LAP)Citation5) produced in a ribosome-mediated pathway. It is produced by Streptomyces sp. TP-A0584, and induces secondary metabolism and cell differentiation in actinomycetes.Citation6) It contains oxazole, methyloxazole, and thiazole rings derived from serine, threonine, cysteine; and dehydroalanine derived from serine (Fig. (A)).Citation7)
Fig. 1. Chemical structure and biosynthetic gene organization of goadsporin.
Note: (A) Chemical structure of goadsporin. (B) Gene organization of goadsporin biosynthetic Gene cluster in Streptomyces sp. TP-A0584. The expression vectors of godI and godR were constructed in this study.
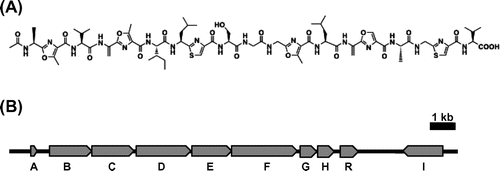
Studies on goadsporin biosynthesis have revealed that goadsporin is synthesized by the ribosome and is post-translationally modified forming heterocyclic rings and dehydroalanines. The structural gene godA encodes 49 amino acids and contains a leader peptide consisting of 30 residues at the N-terminus. godD, godE, godF, godG, and godH are involved in post-translational modification. Based on their amino acid sequences, these genes have been predicted to be involved in various parts of the goadsporin biosynthetic pathway: godD and godE are required to form the heterocyclic ring, godF and godG form dehydroalanine, and godH is involved in N-terminal acetylation (Fig. (B)).
Besides the enzyme coding genes for mature peptide production, there are a pathway-specific regulator gene (godR) and a self-resistance gene (godI) inside the cluster. godR contains a C-terminal helix-turn-helix DNA binding motif, suggesting that it is a pathway specific regulator (SARP, Streptomyces antibiotic regulatory protein)Citation8) for goadsporin biosynthesis. godI is responsible for goadsporin self-resistance and shows 81.6% amino acid sequence identity to the signal recognition particle (SRP) of Streptomyces lividans TK21. SRP is a universally conserved ribonucleoprotein complex that binds to targeting sequences in nascent secretory and membrane proteins.Citation9) We have found that godI is responsible for self-resistance to goadsporin. godI conferred goadsporin resistance to S. lividans over 30 μg/mL, and a godI disruptant in Streptomyces sp. TP-A0584 does not grow on a goadsporin-containing plate,Citation10) but the details of the mechanism of self-resistance to goadsporin is unknown.
Several integrating type vectors that contain the int gene, which encodes an actinophage integrase, have been constructed for the Streptomyces genus. When the vector contains both the int gene and phage attachment site attP, the vector can be integrated into the host strain chromosome by integrase-mediated homologous recombination between attP and attB, the bacterial attachment site in chromosomal DNA. We have constructed two integrating vectors, pTOYAMAcosCitation3) and pTYM1, and confirmed that they are stable in the genome. pTOYAMAcos contains an int gene derived from the phiC31 actinophage and a tsr (thiostreptone resistance) gene isolated from Streptomyces azureus.Citation11) pTYM1 contains the int and neo (aphII, aminoglycoside phosphotransferase) genes, derived from a TG1Citation12) actinophage and Tn5,Citation13) respectively. Thus, pTOYAMAcos and pTYM1 can co-exist in one cell. We have cloned the goadsporin biosynthetic gene cluster using pTOYAMAcos, and heterologous goadsporin production has been successful with S. lividans as surrogate host.Citation10)
Here, we constructed a recombinant strain harboring two and three copies of the goadsporin biosynthetic gene cluster with pTYM1 and pTOYAMAcos. Below, we discuss the effects of gene dosage on goadsporin production per culture volume.
Materials and methods
Bacterial strains, plasmids, and growth conditions
Streptomyces sp. TP-A0584 is a goadsporin-producing strain.Citation6) Streptomyces lividans TK23 was used as heterologous expression host. A-3M,Citation6) a goadsporin production medium, was used for liquid cultures. V-22Citation6) was the seed culture medium used for the Streptomyces strains. Actinomycetes-Escherichia coli bifunctional vectors pTYM1Citation10) and pTOYAMAcosCitation3) were used for the recombination of the S. lividans and Streptomyces sp. TP-A0584 genomes. pGSBC1Citation10) is a cosmid clone selected from the pTOYAMAcos-based genomic library of Streptomyces sp. TP-A0584 that contains the entire goadsporin biosynthetic gene cluster. pGODICitation10) was used for godI expression. E. coli S17-1 was used in transconjugation of integration vectors (Table ).
Table 1. Plasmids used in this study.
Construction of pTYM1-GSBC, pTYM1ep, pGODR, and pGODI
pTYM1-GSBC. pGSBC1 was digested with ClaI and HindIII, and the resulting 6-kb fragment was cloned into pBluescriptII KS+(Stratagene, La Jolla, CA). The plasmid was digested with KpnI and HindIII, and the resulting 6-kb fragment was cloned into the KpnI and HindIII sites of pTYM1 to generate pTYM1-GSB6 k. pGSBC1 was digested with HindIII, and the resulting 14-kb fragment was cloned into the HindIII site of pTYM1-GSB6 k to construct pTYM1-GSBC.
pTYM1ep. the ermE* promoter gene was subcloned as a PCR-generated DNA fragment. Two oligonucleotide primers, 5′-CGCCAATTGCAGGTCCAGGAAGGGGACGTC-3′ and 5′-GCTGAATTCTACCAACCGGCACGATTGTCC-3′, were used to generate the 218-bp ermE* promoter DNA fragment. The underlined letters indicate the MfeI and EcoRI restriction enzyme sites. pM801 was used as template for PCR. The PCR-generated product facilitated further subcloning. The ermE* promoter-containing fragment was cloned into the EcoRI sites of pTYM1 to generate pTYM1ep. pTYM1ep contains a constitutively active ermE* promoter upstream of the multiple cloning site region.
pGODR and pGODI. To construct the godR expression vector pGODR, godR fragment was generated by PCR with pGSBC1 as template. A 0.7-kb fragment was amplified by the use of oligonucleotide primers 5′-CGCCATATGCGTGAAGGCAGCATCATAGCT-3′ and 5′-GCGAAGCTTCAATCGCTATTTAGCGGCGAA-3′. The underlined letters indicate NdeI and HindIII restriction enzyme sites. The resulting fragment was cloned into the NdeI and HindIII sites of pTYM1ep to yield pGODR. The construction of the godI expression vector, pGODI, is described in a previous paper.Citation10)
Quantification of goadsporin by HPLC
Goadsporin was detected by high-performance liquid chromatography (HPLC). Each strain was inoculated into a 500-mL K-1 flask containing 100 mL of V-22 medium. After incubation at 30 °C for 2 d on a rotary shaker at 200 rpm, 5-mL portions of the seed culture were transferred into 500-mL K-1 flasks containing 100 mL of A-3 M medium. Fermentation was done at 30 °C with the same rotary shaker. The culture broth was extracted with an equal volume of n-butanol. The n-butanol was evaporated, and the resulting residue was reconstituted in methanol to prepare samples for HPLC analysis by means of an HP1090 (Hewlett Packard, Palo Alto, CA) system equipped with a C18 Rainin microsorb column (3 μm, 4.6 i.d. × 100 mm, Rainin instrument LLC, Woburn, MA). HPLC analysis was done at 30 °C with acetonitrile-0.15% KH2PO4 (pH 3.5) solvent at a flow rate of 1.2 mL/min. Peaks were detected at 254 nm. The gradient diagram is shown in Fig. . Goadsporin was identified based on its retention time and UV spectrum. Authentic sample of goadsporin was prepared as described in a previous paper.Citation10)
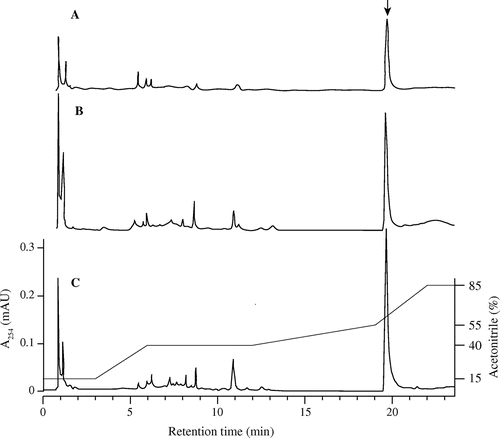
Fig. 2. HPLC profiles of extracts from the culture broth of goadsporin-producing strains.
Note: (A) Streptomyces sp. TP-A0584 wild-type strain. (B) Strain TP-A0584 (pGSBC1), harboring a duplicate set of goadsporin biosynthetic gene clusters in the genome. C, Strain TP-A0584 (pGSBC1 and pTYM1-GSBC) harboring a triplicate set of goadsporin biosynthetic gene clusters in the genome. The arrow in A corresponds to goadsporin. Sample preparation and HPLC conditions are described in “Materials and Methods.” Peaks were detected at 254 nm, the chromatographs were drawn to the same scale, and the gradient diagram is shown on the right-hand scale.
Results
Construction of recombinant strains containing two or three copies of the goadsporin biosynthetic gene cluster
The entire goadsporin biosynthetic gene cluster was duplicated and triplicated in a genome of the original producing strain, TP-A0584, and the production of goadsporin was investigated. pGSBC1 and pTYM1-GSBC contain the entire goadsporin biosynthetic gene cluster and actinomycete genome integration modules that consist of integration genes isolated from actinophages phiC31Citation14) and TG1.Citation15) Each integrase recognizes a different attB site in a genome, enabling the two plasmids to integrate into a single genome. After transconjugation, site-specific integrases phiC31 and TG1 catalyze unidirectional recombination between the vector attP and the chromosomal attB, and single integrated recombination occurs at the attB site.Citation16) Usually, the target attB sites for the phiC31 and TG1 attB integrases are located in a pirin-like protein and aminotransferase,Citation16) and the attB sites in the S. lividans genome have been reported.Citation16,17) In the draft genome of Streptomyces sp. TP-A0584, we also found single attB sites for both integrases. The attB sequence of phiC31 in Streptomyces ambofaciens is 5′-TGACGGTCTCGAAGCCGCGGTGCGGGTGCCAGGGCGTGCCCTTGGGCTCCCCGGGCGCGTACTCCACCTCACCCATCTGGTCCA-3′,Citation18) and a corresponding sequence exists in the draft genome sequence of strain TP-A0584, 5′-TGACGGTCTCGAAGCCGCGGTGCGGGTGCCAGGGGGTGCCCTTGGGCTCGCCCGGCGCGTACTCCACCTC-3′.
The attB site of TG1 in Streptomyces avermitilis is 5′-GATCAGCTCCGCGGGCAAGACCTTCTCCTTCACGGGGTGGAAGGTC-3′,Citation19) and a corresponding sequence exists in the draft genome sequence of strain TP-A0584, 5′-GACCATCTCGTCGGCCGGGAAGACGTTCTCGTTCACCGGCTGGAAGGTG-3′.
To construct a strain harboring two copies of the goadsporin biosynthetic gene cluster, pGSBC1 was introduced into Streptomyces sp. TP-A0584 to produce strain TP-A0584 (pGSBC1). Then, to construct a strain harboring three copies of the goadsporin biosynthetic gene cluster, pTYM1-GSBC was introduced into strain TP-A0584 (pGSBC1) to yield strain TP-A0584 (pGSBC1 and pTYM1-GSBC). The growth of the transformants did not show any significant difference.
Quantification of goadsporin from recombinant strains
Goadsporin production was quantified by HPLC. Culture broths of the strains were extracted with equal volumes of n-butanol. Five-milliliter aliquots were taken from culture broths of TP-A0584, TP-A0584 (pGSBC1), and TP-A0584 (pGSBC1 and pTYM1-GSBC). The aliquots were extracted with 5 mL of n-butanol. The extracts were analyzed by HPLC (Fig. ), and the amount of goadsporin produced by each strain was determined. Goadsporin production is plotted in Fig. . All the strains reached maximum production on day 7. The amounts of goadsporin produced on day 7 showed an average of 126.3 mg/L in the wild-type strain, an average of 183.4 mg/L in TP-A0584 (pGSBC1), and an average of 283.4 mg/L in TP-A0584 (pGSBC1 and pTYM1-GSBC). The recombinant strains harboring two or three copies of the biosynthetic gene cluster produced 1.46-fold or 2.25-fold more goadsporin than the wild-type strain.
Fig. 3. Amounts of goadsporin produced by Streptomyces sp. TP-A0584 by the duplicate and triplicate gene cluster containing strains.
Note: Circles, Streptomyces sp. TP-A0584; triangles, strain TP-A0584 (pGSBC1); squares, strain TP-A0584 (pGSBC1 and pTYM1-GSBC). The concentrations of goadsporin are average values for two independent experiments, TP-A0584 and TP-A0584 (pGSBC1) and three independent experiments, TP-A0584 (pGSBC1 and pTYM1-GSBC).
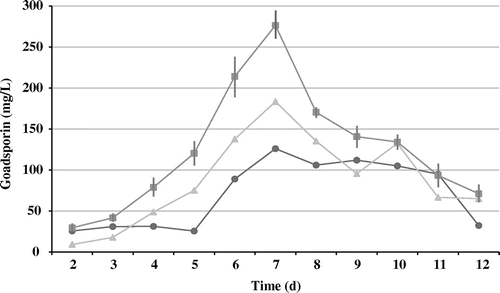
Heterologous production of goadsporin in a surrogate host
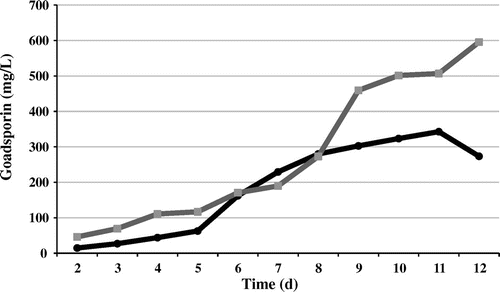
We investigated the effect of gene dosage on a surrogate host, Streptomyces lividans TK23, by introducing pGSBC1 and pTYM1-GSBC into S. lividans and constructing recombinant strains S. lividans (pGSBC1) and S. lividans (pGSBC1 and pTYM1-GSBC), which harbored one or two copies of the goadsporin biosynthetic gene cluster. The aliquots extracted with n-butanol were analyzed by HPLC. Goadsporin production is shown in Fig. . The amounts of goadsporin produced on day 11 in S. lividans (pGSBC1) and day 12 in S. lividans (pGSBC1 and pTYM1-GSBC) showed an average of 342.7 mg/L and an average of 595.3 mg/L, respectively. The duplicate strain (pGSBC1 and pTYM1-GSBC) produced 1.74-fold more goadsporin than the single-copy strain (pGSBC1).
Transcriptional and self-resistance effects on goadsporin production
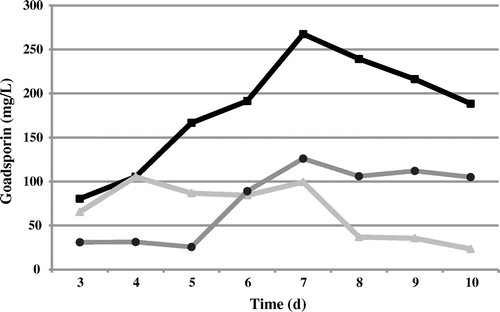
We examined the effects of a transcriptional regulator and a self-resistance gene on goadsporin productivity. godR, which is located in the goadsporin biosynthetic gene cluster, has a protein motif of LuxR-type DNA-binding domain, suggesting that godR is a SARPCitation8) in goadsporin biosynthesis. godI, a homolog of ffh, was predicted to be a goadsporin self-resistance gene.Citation10) godR and godI were separately cloned into pTYM1 under the ermE* promoter, and integrated into the TP-A0584 chromosomal DNA. The amount of goadsporin produced is shown in Fig. . The godR and godI transformants reached maximum production on day 7 and day 4, respectively. The godR and godI transformants produced an average of 267.5 mg/L and an average of 105.3 mg/L on days 7 and 4, respectively. Compared to the wild-type strain, the amount of goadsporin produced by the godR transformant on day 7 was 2.12-fold higher, and the godI transformant on day 7 was 0.79-fold lower. However, when we made this comparison for day 4, the godI transformant produced 3.36-fold more strongly than the wild-type strain. These results suggest that godR, a SARP transcriptional regulator homolog, is an activator and that the overexpression thus leads to high production of goadsporin. Overexpression of the self-resistance gene, godI made production peak earlier, but did not increase the level of production.
Fig. 5. Transcriptional and self-resistance effects on goadsporin production.
Note: Circles, Streptomyces sp. TP-A0584 (control); triangles, godI constitutive expression strain of Streptomyces sp. TP-A0584; squares, godR constitutive expression strain of Streptomyces sp. TP-A0584. The concentrations of goadsporin are average values for two independent experiments.
Discussion
In this study, we examined the gene dosage effects of a biosynthetic gene cluster involved in the production of a secondary metabolite. Although the recombinant techniques used to deal with the biosynthetic gene cluster of a secondary metabolite spanning over 20 kb are cumbersome and complicated, pTYM1 and pTOYAMAcos co-exist stably on one Streptomyces chromosome, enabling the accurate estimation of gene dosage effects in Streptomyces cells harboring up to three copies. In the TP-A0584 strains harboring duplicate and triplicate goadsporin biosynthetic gene clusters, goadsporin production increased by 1.46- and 2.25-fold, respectively, as compared to production in the wild-type strain on day 7. In comparison to heterologous strains harboring one and two copies of the gene cluster, goadsporin production increased 1.74-fold in the strain containing two copies. These results suggest that the biosynthetic gene cluster duplication procedure can be applied to improve goadsporin production.
Gene dosage effects as to goadsporin production were also observed in a surrogate host, S. lividans. The S. lividans transformants produced more goadsporin than the native producer. Goadsporin production in the native strains decreased after day 7, whereas in the surrogate host, it continued to increase through day 11. Although the details of the mechanism remain unknown, these results suggest that some goadsporin degradation systems exist in the native strain.
To determine the effects of SARP (godR) and a self-resistance gene (godI) on goadsporin production, we used a constitutive promoter, ermE* to express both genes. The differences between the production patterns of the cluster-integration strain and the godR transformant depended on the differences between their promoters. Enhancing the godI self-resistance gene was not effective in an attempt to increase goadsporin production, but the onset of production was accelerated, suggesting that transcription of the ermE* promoter is more efficient than that of the native godI promoter at the early stage of cell growth, and that the resistance mechanism of cells is critical for the onset of secondary metabolism. Our results also indicate that the timing of SARP expression is critical for increases in the production of secondary metabolites.
Supplemental material
The supplemental material for this paper is available at http://dx.doi.org/10.1080/09168451.2014.885824.
Supplementary Fig. 1
Download MS Power Point (106 KB)Supplementary Fig. 1 caption
Download MS Word (28.6 KB)Funding
This work was supported in part by grants-in-aid from the New Energy and Industrial Technology Development Organization (NEDO) of Japan (to H. O.), Institution of Fermentation, Osaka (to S. A., T. O., and H. O.), and a JSPS KAKENHI (grant number 25108707 to H. O.).
References
- Murakami T, Burian J, Yanai K, Bibb MJ, Thompson CJ. A system for the targeted amplification of bacterial gene clusters multiplies antibiotic yield in Streptomyces coelicolor. Proc. Natl. Acad. Sci. USA. 2011;108:16020–16025.10.1073/pnas.1108124108
- Hatanaka T, Onaka H, Arima J, Uraji M, Uesugi Y, Usuki H, Nishimoto Y, Iwabuchi M. pTONA5: a hyperexpression vector in Streptomycetes. Protein Expr. Purif. 2008;62:244–248.10.1016/j.pep.2008.09.001
- Onaka H, Taniguchi S, Ikeda H, Igarashi Y, Furumai T. pTOYAMAcos, pTYM18, and pTYM19, actinomycete-Escherichia coli integrating vectors for heterologous gene expression. J. Antibiot. 2003;56:950–956.10.7164/antibiotics.56.950
- Onaka H, Tanifguchi S, Igarashi Y, Furumai T. Cloning of the staurosporine biosynthetic gene cluster from Streptomyces sp. TP-A0274 and its heterologous expression in Streptomyces lividans. J. Antibiot. 2002;55:1063–1071.10.7164/antibiotics.55.1063
- Arnison PG, Bibb MJ, Bierbaum G, Bowers AA, Bugni TS, et al. Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 2012;30:108–160.10.1039/c2np20085f
- Onaka H, Tabata H, Igarashi Y, Sato Y, Furumai T. Goadsporin, a chemical substance which promotes secondary metabolism and morphogenesis in streptomycetes. I. Purification and characterization. J. Antibiot. 2001;54:1036–1044.10.7164/antibiotics.54.1036
- Igarashi Y, Kan Y, Fujii K, Fujita T, Harada K, Naoki H, Tabata H, Onaka H, Furumai T. Goadsporin, a chemical substance which promotes secondary metabolism and Morphogenesis in streptomycetes. II. Structure determination. J. Antibiot. 2001;54:1045–1053.10.7164/antibiotics.54.1045
- Wietzorrek A, Bibb M. A novel family of proteins that regulates antibiotic production in streptomycetes appears to contain an OmpR-like DNA-binding fold. Mol. Microbiol. 1997;25:1181–1184.10.1046/j.1365-2958.1997.5421903.x
- Valent QA. Signal recognition particle mediated protein targeting in Escherichia coli. Antonie Van Leeuwenhoek. 2001;79:17–31.10.1023/A:1010256109582
- Onaka H, Nakaho M, Hayashi K, Igarashi Y, Furumai T. Cloning and characterization of the goadsporin biosynthetic gene cluster from Streptomyces sp. TP-A0584. Microbiology. 2005;151:3923–3933.10.1099/mic.0.28420-0
- Thompson CJ, Kieser T, Ward JM, Hopwood DA. Physical analysis of antibiotic-resistance genes from Streptomyces and their use in vector construction. Gene. 1982;20:51–62.10.1016/0378-1119(82)90086-5
- Hirano N, Muroi T, Kihara Y, Kobayashi R, Takahashi H, Haruki M. Site-specific recombination system based on actinophage TG1 integrase for gene integration into bacterial genomes. Appl. Microbiol. Biotechnol. 2011;89:1877–1884.10.1007/s00253-010-3003-7
- Beck E, Ludwig G, Auerswald EA, Reiss B, Schaller H. Nucleotide sequence and exact localization of the neomycin phosphotransferase gene from transposon Tn5. Gene. 1982;19:327–336.10.1016/0378-1119(82)90023-3
- Thorpe HM, Smith MC. In vitro site-specific integration of bacteriophage DNA catalyzed by a recombinase of the resolvase/invertase family. Proc. Natl. Acad. Sci. USA. 1998;95:5505–5510.10.1073/pnas.95.10.5505
- Morita K, Yamamoto T, Fusada N, Komatsu M, Ikeda H, Hirano N, Takahashi H. The site-specific recombination system of actinophage TG1. FEMS Microbiol. Lett. 2009;297:234–240.10.1111/fml.2009.297.issue-2
- Baltz RH. Streptomyces temperate bacteriophage integration systems for stable genetic engineering of actinomycetes (and other organisms). J. Ind. Microbiol. Biotechnol. 2012;39:661–672.10.1007/s10295-011-1069-6
- Rausch H, Lehmann M. Structural analysis of the actinophage phi C31 attachment site. Nucleic Acids Res. 1991;19:5187–5189.10.1093/nar/19.19.5187
- Kuhstoss S, Rao RN. Analysis of the integration function of the streptomycete bacteriophage phi C31. J. Mol. Biol. 1991;222:897–908.10.1016/0022-2836(91)90584-S
- Morita K, Morimura K, Fusada N, Komatsu M, Ikeda H, Hirano N, Takahashi H. Site-specific genome integration in alphaproteobacteria mediated by TG1 integrase. Appl. Microbiol. Biotechnol. 2012;93:295–304.10.1007/s00253-011-3545-3