Abstract
A yam (Dioscorea opposita Thunb) class IV chitinase, whose genomic DNA was cloned by Mitsunaga et al. (2004), was produced by the recombinant Pichia pastoris X-33 in high yields such as 66 mg/L of culture medium. The chitinase was purified by column chromatography after Endoglycosidase H treatment and then characterized. It showed properties similar to the original chitinase E purified from the yam tuber reported by Arakane et al. (2000). This Pichia-produced chitinase also showed strong lytic activity against Fusarium oxysporum and Phytophthora nicotianae, wide pH and thermal stability, optimum activity at higher temperature such as 70 °C, and high substrate affinity, indicating that one can use this Pichia-produced yam chitinase as a bio-control agent.
Graphical Abstract
Yam clsss IV chitinase produced by recombinant Pichia pastoris X-33 showed lytic activity against fungal pathogens as well as the original yam chitinase E.
Chitinases [EC 3.2.1.14] are diversified in biological functions depending on the source. Chitinases, which hydrolyze chitin, occur in a wide range of organisms, including viruses, bacteria, fungi, insects, higher plants, and animals.Citation1) Bacterial chitinases play roles in nutrition and parasitism, while fungal chitinases have autolytic, nutritional, and morphogenetic roles. Viral chitinases are involved in pathogenesis.Citation2) Invertebrate chitinases play roles in digestion. In insects and crustaceans, chitinases are associated with ecdysis through degradation of old cuticle.Citation3) Some plant chitinases are involved in plant defense mechanisms against fungal and bacterial pathogens through lytic action against chitin and peptidoglycan (lysozymic action), respectively,Citation4,5) and show in vitro antifungal properties against several phytopathogenic fungi, inhibiting both spore germination and hyphal growth either alone or synergistically with other pathogenesis-related proteins;Citation6–8) while other chitinases and chitinase isoforms play important roles in the growth and development of plants, as in embryonic development, pollination, and sexual reproduction.Citation9–11) Some plant chitinases are reported to play roles in the protection of plants against environmental stress.Citation11)
Chitinases have immense potential for biotechnological applications, as in preparing pharmaceutically important chitooligosaccharides and N-acetyl d-glucosamine, which are promising antibacterial agents, lysozyme-inducing elicitors, and immunoenhancers; the production of single-cell proteins; protoplast isolation from fungi and yeasts; the development of bio-control agents for pests and pathogens, and disease resistant transgenic plants; the control of mosquitoes to interfere with or block the transmission of diseases such as yellow fever, dengue, and malaria; and chitinous waste management.Citation12–14)
An isozyme of plant chitinase, chitinase E, purified from yam tuber, was found to have strong lytic activity against fungal pathogens, indicating its possible application as a bio-control agent.Citation15,16) Hence the genomic DNA of yam class IV chitinase was cloned along with its tentative vacuolar targeting signal (VTS) at the C-terminal on the basis of the partial amino acid sequences of yam chitinase E.Citation17) In the present study, we made an attempt to produce this yam chitinase in high yield by means of the recombinant Pichia carrying its gene. A major advantage of Pichia pastoris over a bacterial expression system is that the yeast has the potential to perform many of the post-translational modifications typically associated with higher eukaryotes, such as the processing of signal sequences, folding, disulfide bridge formation, certain types of lipid addition, and O- and N-linked glycosylation.Citation18) Hence, we investigated the possibility of using this yam chitinase as a bio-control agent as well as yam chitinase E.
Materials and methods
Plasmids, vectors, and host strains
pBluescript SK+ (Stratagene Inc.) and pDrive (Qiagen K.K.) were used as cloning vectors, and were transformed into the Escherichia coli JM 109 as host. Vector pPICZαA (Invitrogen Corp.) was used as expression vector for the chitinase gene. The methylotrophic yeast P. pastoris X-33 (Invitrogen Corp.) was used as expression host for the production of recombinant chitinase.
Cloning of the yam class IV chitinase gene without intron and VTS
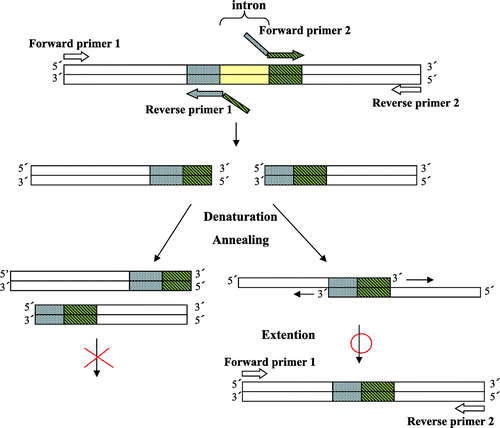
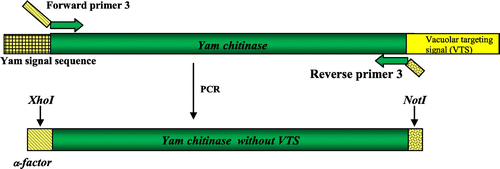
Molecular cloning of a genomic DNA fragment containing the full-length open reading frame of the family 19 yam class IV chitinase gene (accession no. AB102714), which included one intron 124 bp long, was done previously.Citation17) First, the intron was removed by three consecutive rounds of polymerase chain reaction (PCR) with the following primer pairs: forward primer 1 and reverse primer 1, forward primer 2 and reverse primer 2, and forward primer 1 and reverse primer 2 (Table ), respectively, as summarized in Fig. . Then removal of the C-terminal VTS of 24 bp and replacement of the N-terminal signal sequence of yam chitinase with α-factor of the expression vector pPICZαA were done as shown in Fig. by PCR with forward primer 3 and reverse primer 3 (Table ). These primers contained the site for XhoI and a partial sequence for α-factor, respectively, and a NotI digestion site along with stop codon. PCR amplification of the gene was done with Elongase Enzyme Mix (Life Technologies Corp.). Thus, we obtained the yam class IV chitinase gene without intron and VTS.
Table 1. Oligonucleotide primer sequences used in PCR.
Construction of expression vector and recombinant P. pastoris X-33
Construction of a recombinant expression vector (pPICZαA) containing the functional yam chitinase gene and electroporation of the expression cassette into competent P. pastoris X-33 were done by the method of Saito et al.Citation19)
Large-scale expression of recombinant Pichia strain
A 3 mL preculture of recombinant P. pastoris was developed in YPDM + ZeocineTM medium (1% yeast extract, 2% peptone, 2% dextrose, 0.5% methanol, and 100 μg/mL of ZeocinTM) in a 50 mL test tube at 30 °C for 48 h at 140 strokes/min in a shaker water bath. For the production of chitinase, the preculture was transferred to 200 mL of SM medium in a 500-mL flask and incubated for 8 d at 30 °C at 180 rpm in a rotary shaker incubator (Bio-shaker BR300LF; Taitec Corp.). The SM medium contained 0.2% (NH4)2SO4, 1% KH2PO4, 0.01% CaCl2, 0.2% MgSO4/7H2O, 0.2% KCl, 0.01% NaCl, 0.01% ZnSO4/7H2O, 0.0005% CuSO4/5H2O, 0.01% FeCl3/6H2O, 10−6% biotin, 5 × 10−5% thiamine hydrochloride, 5 × 10−5% pyridoxine hydrochloride, 5 × 10−5% sodium pantothenate, 2 × 10−3% inositol, and 100 mm sodium phosphate buffer, pH 7.5. For chitinase induction, methanol was added to the culture medium every 24 h as carbon source to maintain a final concentration of 0.5%. Chitinase was harvested from culture supernatant by centrifugation at 8000 g for 15 min at 4 °C. The harvested chitinase in culture supernatant was desalted by dialysis against 10 mm sodium phosphate buffer, pH 8.0, in a cellulose tube with a cut-out molecular mass of 10,000 Da. Activity assay and activity staining were performed before and after dialysis.
Purification of P. pastoris-produced yam chitinase
The crude enzyme harvested in the supernatant of the culture medium was deglycosylated by treatment with Endoglycosidase H (Endo H) (New England Biolabs Inc.) at a rate of 4 μL of Endo H (500 unit/μL) for 400 mL of culture supernatant with incubation at 27 °C for 24 h. The flocculated carbohydrate moieties released from the Pichia-produced chitinase by the action of Endo H were removed by centrifugation at 8000 g over 20 min. The treated enzyme was applied to an anion-exchange resin, DEAE-Toyopearl 650(M) (Tosoh Corp.), which was packed in a glass column (φ2 × 50 cm) and pre-equilibrated with 10 mm sodium phosphate buffer, pH 8.0. The column was washed with the same buffer to elute the unbound proteins, and then the bound chitinase was eluted with a linear gradient of NaCl from 0 to 0.5 m in the same buffer. The selected active fractions were applied to Fractogel EMD DEAE 650(M) (Merck KGaA) in a glass column (φ1.2 × 20 cm), and elution was done with an NaCl linear gradient from 0 to 0.5 m in the same buffer. The active fractions were again subjected to DEAE-Toyopearl 650(M) chromatography. The active fractions from the second DEAE-Toyopearl 650(M) were dialyzed against 50 mm sodium phosphate buffer, pH 8.0, containing 0.2 m sodium chloride and put on a Sephacryl S-100 (GE Healthcare Japan Corp.) column (φ2 × 95 cm) equilibrated with the same buffer. The active fractions were pooled, dialyzed against 10 mm sodium phosphate buffer, pH 8.0, and chromatographed on a DEAE-Toyopearl 650(M) column (φ1 × 30 cm) with a sodium chloride linear gradient from 0 to 0.3 m in the same buffer.
Protein measurement
To monitor proteins during chromatographic separation, absorbance was detected at 280 nm. The protein concentration was measured by the method of Lowry et al.Citation20) with bovine serum albumin as standard.
Enzyme assay
Chitinase activity was measured with glycolchitin as substrate. For purification, 5–20 μL of chitinase solution was added to 0.5 mL of 0.05% (w/v) glycolchitin dissolved in 50 mm sodium phosphate buffer, pH 8.0, and this was incubated at 32 °C for 30 min. The reducing end group produced was measured colorimetrically at 420 nm with a ferri-ferrocyanide reagent by the method of Imoto and Yagishita.Citation21) Britton-Robinson buffer (pH 2.0–12.0)Citation22) was also used to determine the pH optimum and stability. The enzymatic reaction proceeded linearly with time to 20% completion of the reaction with glycolchitin.
For kinetic analysis, the purified chitinase (final concentration, 50 nm) was incubated with 0.025–0.4 mg/mL of glycolchitin in 50 mm sodium phosphate buffer, pH 8.0, at 25 °C. The initial velocity was calculated from the difference in the absorbance at 420 nm between sample and control experiments assuming a relationship of 1 ΔA420 = 0.22 mm N-acetylchitooligosaccharides. The kinetic parameters such as Km and Vmax (= kcat × E0) were obtained by double reciprocal plots by the method of Lineweaver and Burk.Citation23)
To determine stability to dryness, 100 μL of the purified chitinase (8 μg) was kept in an open Eppendorf tube in an incubator at 25 °C for 7 d. The control sample was kept closed at 4 °C. After 7 d, the residual dry mass at the bottom of the tube was recovered by solubilization with 100 μL of distilled water. Then the remaining activity of the dried chitinase was measured and compared with that of the control.
To determine lytic activity, the purified chitinase was incubated with mycelia of fungal pathogen in 25 mm sodium phosphate buffer, pH 8.0, containing 3% Zymolyase 20T (Seikagaku Corp.), whose main component is β-1,3-glucanase, and 0.4% magnesium sulfate, which was used to stabilize the protoplasts, at 30 °C for 8 h. The protoplasts released were counted under a light microscope.
Polyacrylamide gel electrophoresis (PAGE) and activity staining
SDS-PAGE was performed with a 10% polyacrylamide slab gel containing 0.1% sodium dodecyl sulfate (SDS) by the method of Laemmli.Citation24) Molecular weight was measured by SDS-PAGE (10% gel) with standard marker proteins (Takara Bio Inc.) including rabbit muscle phosphorylase b (97.2 kDa), bovine serum albumin (66.4 kDa), hen egg white ovalbumin (44.3 kDa), bovine carbonic anhydrase (29 kDa), soybean trypsin inhibitor (20.1 kDa), and hen egg white lysozyme (14.3 kDa). Native PAGE was done with a 7.5% polyacrylamide gel pH 9.5 by the method of Davis.Citation25) Chitinase activity staining was done with 0.02% fluorescent brightener 28 (Sigma) by the method of Koga et al.Citation26) After electrophoresis, the proteins in the gel (85 × 45 × 1 mm) were transblotted electrophoretically on to a similar polyacrylamide gel containing 0.01% glycolchitin as substrate with a semi-dry type blotting apparatus (AE-6670P/N; Atto Corp.) at 90 mA for 30 min after SDS-PAGE or for 10 min after native PAGE. The chitinase active band was observed as a black band under UV irradiation.
N-Terminal sequence analysis
The purified chitinase was checked as to its N-terminal sequence by the automated Edman degradation method. For this purpose, the chitinase was treated with pyroglutamate amino peptidase and then separated by SDS-PAGE followed by transfer to a PVDF membrane (Immobilon P; Millipore Corp.) by electroblotting and stained with Coomassie Brilliant Blue R-250. Then the excised protein bands were subjected to N-terminal sequence analysis with PPSQ-21A (Shimadzu Corp.).
Results
Sequence analysis of the yam class IV chitinase gene introduced into recombinant P. pastoris X-33
The yam chitinase gene carried by recombinant P. pastoris X-33 was sequenced and compared with the complete nucleotide sequence of the class IV yam chitinase by homology alignment. The results indicated precise deletion of both intron and VTS and in-frame fusion of the α-factor instead of the signal sequence in recombinant Pichia carrying yam chitinase gene.
Purification of the chitinase from recombinant P. pastoris culture
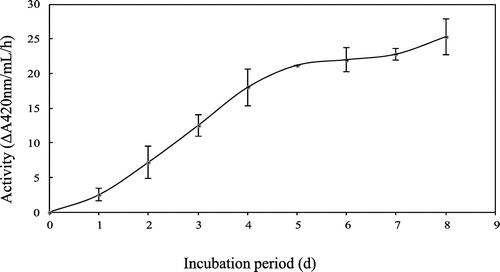
Chitinase secretion in the culture medium almost reached a plateau after 5 d of incubation (Fig. ). The crude enzyme, extracted from culture medium by centrifugation, showed five main active bands by activity staining after SDS-PAGE with molecular masses of about 58, 48, 38, 32, and 28 kDa (Fig. (A)). After deglycosylation by treatment with Endo H, the pattern of the chitinase active band shifted to smaller molecular masses, at 32 and 28 kDa. In order to obtain 32 kDa-chitinase, which is similar in molecular size to yam chitinase E (33.5 kDa), indicating its possible application as a bio-control agent,Citation15,16) the following course of purification was applied. The deglycosylated crude enzyme after dialysis against 10 mm sodium phosphate buffer, pH 8.0, was applied sequentially to DEAE-Toyopearl 650(M), Fractogel EMD DEAE 650(M), DEAE-Toyopearl 650(M), Sephacryl S-100, and DEAE-Toyopearl 650(M) resins. The active peaks of the first chromatographic separation on DEAE-Toyopearl 650(M) were obtained at sodium chloride conductivities between 7.5 and 18 mS (Fig. (A)). The active fractions, indicated by the horizontal bar, were combined and applied to Fractogel EMD DEAE 650(M), and the active fractions were obtained between 15 and 27 mS of sodium chloride conductivity. These were combined and applied again to DEAE-Toyopearl 650(M), followed by Sephacryl S-100 (Fig. (B)). The active fractions of small molecular sizes of about 32 kDa were pooled by gel filtration chromatography and eluted again by DEAE-Toyopearl 650(M) with a gradient of 0–0.3 m sodium chloride (Fig. (C)). Purity analysis by both SDS-PAGE and native PAGE showed a single homogeneous band with a molecular mass of 32 kDa (Fig. ). The quantitative results for purification steps are presented in Table . A 3% yield with 10-fold overall purification was achieved at the final step with a specific activity of 374 ΔA420/h/mg.
Fig. 4. SDS-PAGE of culture medium of recombinant P. pastoris X-33 before and after treatment with Endoglycosidase H (Endo H).
Notes: Panels: (A) Chitinase activity staining; (B) protein staining. Lanes: 1, before treatment with Endo H; 2, after treatment with Endo H; M, molecular markers.
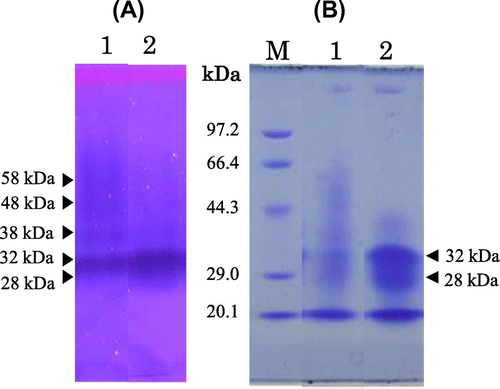
Fig. 5. Purification of yam chitinase from recombinant P. pastoris X-33.
Notes: Panels: (A) The first DEAE-Toyopearl 650(M) chromatography of Endo H-treated crude chitinase; (B) Sephacryl S-100 gel filtration of the selected active fraction obtained by the second round of DEAE-Toyopearl 650(M) chromatography following Fractogel EMD DEAE 650(M); (C) the third round of DEAE-Toyopearl 650(M) chromatography of the selected active fraction obtained by Sephacryl S-100 gel filtration. Active fractions indicated by the horizontal bar were used in the following experiment.
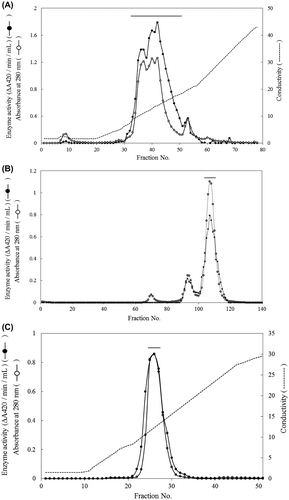
Fig. 6. PAGE of the yam chitinase purified from recombinant P. pastoris X-33.
Notes: Lanes: A, B, and M, SDS-PAGE; C and D, native PAGE; A and D, chitinase activity staining; B, M, and C, protein staining; A, B, C, and D, purified chitinase; M, molecular markers.
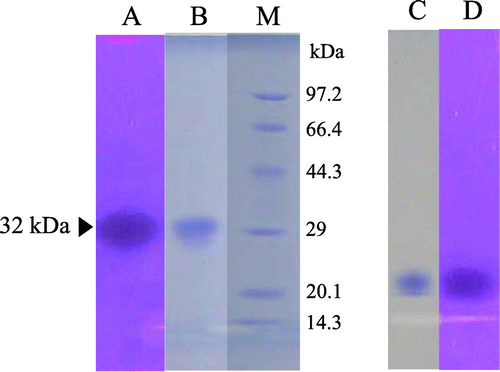
Table 2. Purification of P. pastoris-produced yam chitinase.
N-Terminal amino acid sequencing
Automated Edman degradation of the chitinase produced by recombinant P. pastoris was unsuccessful. When digested with pyroglutamate amino peptidase, the N-terminals were read to be SYD, the same as the amino acid sequence of the introduced yam chitinase from the 21st. This result was obtained repeatedly, suggesting that appearance of the SYD sequence from the 21st amino acid is due to the action of peptidase contaminating in this pyroglutamate amino peptidase.
pH optimum and pH stability of yam chitinase purified from recombinant P. pastoris X-33
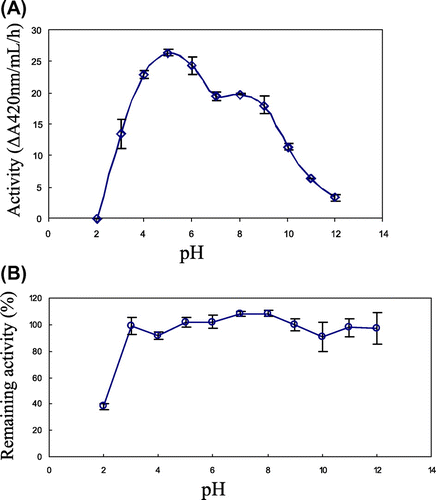
To determine the optimum pH, the activity of the purified chitinase was measured with glycolchitin in Britton-Robinson buffer, pH 2–12, at 32 °C. The result is shown in Fig. (A). Pichia-produced yam chitinase showed highest activity at pH 5 and a second peak at pH 8.0 with comparatively low activity. To determine pH stability, the purified chitinase was treated with Britton-Robinson buffer, pH 2–12, for 12 h at 4 °C. The result is shown in Fig. (B). The Pichia-produced yam chitinase was stable over a wide range of pH from 3 to 12. Even at pH 2, it retained activity at 38%.
Fig. 7. pH optimum and pH stability of yam chitinase purified from recombinant P. pastoris X-33.
Notes: Panels: (A) The optimum pH of the purified chitinase was determined by incubation of 50 nm chitinase with 0.05% glycolchitin in Britton-Robinson buffer, pH 2.0–12.0, at 32 °C for 5–60 min; (B) the pH stability of the purified chitinase was determined by measuring the remaining activity after incubation of 50 nm chitinase in Britton-Robinson buffer, pH 2.0–12.0, at 4 °C for 120 h.
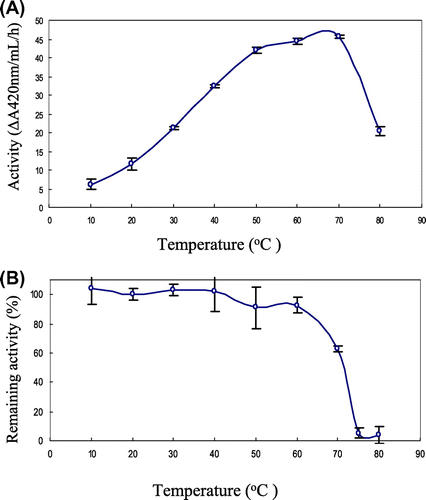
Temperature optimum and thermal stability of yam chitinase purified from recombinant P. pastoris X-33
To determine the optimum temperature, the activity of the purified chitinase was measured with glycolchitin at 10–80 °C in Britton-Robinson buffer, pH 8.0. The result is shown in Fig. (A). Pichia-produced yam chitinase showed highest activity at 70 °C. To determine thermal stability, the purified chitinase was treated at 10–80 °C for 15 min in Britton-Robinson buffer, pH 8.0. The result is shown in Fig. (B). The Pichia-produced yam chitinase was stable at up to 70 °C with remaining activity of more than 60%. The stability with above 90% was observed up to 60 °C, but at 75 °C it retained activity at only 10%.
Fig. 8. Temperature optimum and thermal stability of the yam chitinase purified from recombinant P. pastoris X-33.
Notes: Panels: (A) The optimum temperature of the purified chitinase was measured by incubation of 50 nm chitinase with 0.05% glycolchitin in Britton-Robinson buffer, pH 8.0, for 5–60 min at temperatures of 10–80 °C; (B) the thermal stability of the purified chitinase was determined by measuring the remaining activity after incubation of 50 nm chitinase in Britton-Robinson buffer, pH 8.0, for 15 min at temperature range of 10–80 °C.
Stability of the yam chitinase purified from recombinant P. pastoris X-33 to dryness
To determine stability to dryness, the purified chitinase was exposed to air at 25 °C for 7 d. Remaining activity was measured with glycolchitin and was compared with the control kept closed at 4 °C. The stability of the Pichia-produced yam chitinase was up to 68.4% of the remaining activity.
Kinetic analysis of the yam chitinase purified from recombinant P. pastoris X-33
To investigate the enzymatic action, kinetic analysis was done with glycolchitin as substrate. The Km, kcat, and kcat/Km values were obtained by double reciprocal plot.Citation23) They are shown in Table .
Table 3. Kinetic parameters of P. pastoris-produced yam chitinase and yam chitinase E.
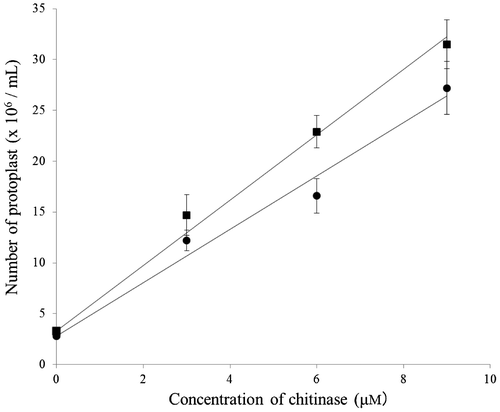
Lytic activity of the yam chitinase purified from recombinant P. pastoris X-33
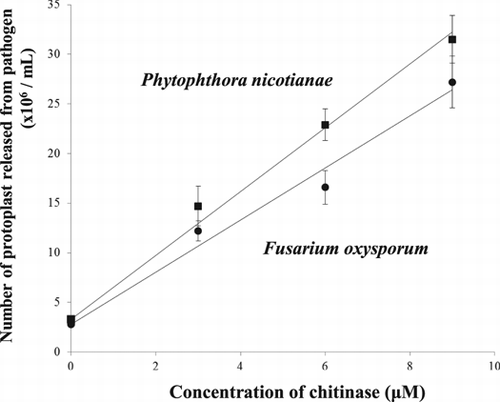
The lytic activity of the purified chitinase against pathogens was investigated by counting the protoplasts released from the pathogen under conditions stabilizing the released protoplasts. As pathogens, Phytophthora nicotianae and Fusarium oxysporum were used. The results are shown in Fig. . In both cases, the numbers of protoplasts released increased depending on the concentration of purified chitinase, indicating that protoplast release was due to the chitinase action. The lytic activity of the Pichia-produced yam chitinase was higher against Phytophthora than against Fusarium.
Fig. 9. Lytic activity of yam chitinase purified from recombinant P. pastoris X-33 against fungal pathogens.
Notes: The lytic activity of the purified chitinase was measured under a light microscope by counting the protoplasts released from mycelia of fungal pathogens such as F. oxysporum (solid circle) and P. nicotianae (solid square) after incubation of 3–9 μm chitinase with 2 mg pathogen mycelium in 0.2 mL of 25 mm sodium phosphate buffer, pH 8.0, containing 3% Zymolyase and 0.4% magnesium sulfate at 30 °C for 8 h.
Discussion
As shown in Fig. , a family 19 Class IV acidic yam chitinase without VTS was produced increasingly with time by recombinant P. pastoris X-33. The results of SDS-PAGE for Pichia-produced chitinase in 5-d culture medium, however, showed several chitinase active bands of various molecular sizes from 28 to 58 kDa (Fig. ). On the other hand, the molecular weight of yam chitinase E purified from yam tuber has been reported to be 33.5 kDa,Citation15) while results for molecular cloning of it indicated that the molecular mass is 28 kDa.Citation17) Therefore, the extra mass over 28 kDa must be due to carbohydrate moiety. After treatment of the culture medium with Endo H, chitinase active bands shifted as expected mainly to 32 kDa, which is similar to the molecular weight of yam chitinase E (33.5 kDa). Regarding chitinase activity toward glycolchitin, no effect of Endo H treatment was observed. After treatment with Endo H, the 32-kDa chitinase was purified by column chromatography (Table ). The specific activity of the purified chitinase was 374ΔA420/h/mg. By this specific activity, the production of chitinase in an 8-d culture medium was calculated to be 66 mg/L. This is considerably high production.
In order to compare the Pichia-produced yam chitinase with yam chitinase E, the purified chitinase was investigated as to optimum activity and stability. The results are summarized and compared with those of yam chitinase E in Table . With respect to physicochemical properties, as shown in Fig. , as to SDS-PAGE and native PAGE, the purified chitinase was an acidic chitinase with molecular mass of 32 kDa. The N-terminal amino acid was not detected from both Pichia-produced yam chitinase and yam chitinase E by the Edman degradation method. With respect to activity, Pichia-produced yam chitinase showed two pH optima, in acidic and alkaline ranges, which is one of the characteristics of yam chitinase E. Both chitinases showed an optimum temperature of 70 °C. On the other hand, the stability of chitinase is important for its application as a bio-control agent. With respect to pH stability, the Pichia-produced yam chitinase showed a wider range of pH than yam chitinase E, and they showed the same thermal stability. Furthermore, the Pichia-produced yam chitinase showed high stability against dryness.
Table 4. Comparative characteristics of P. pastoris-produced yam chitinase and yam chitinase E.
With respect to kinetic behavior, the Pichia-produced yam chitinase was found to be better in terms of substrate affinity due to its lower Km value (0.233) toward glycolchitin than yam chitinase E (Km = 0.518), indicating that the Pichia-produced yam chitinase, with a smaller Km value, had more substrate affinity than yam chitinase E, but kcat (0.200) was lower than that of yam chitinase E (kcat = 0.645). Thus, the overall reaction (kcat/Km = 0.858) of Pichia-produced yam chitinase toward glycolchitin was found to be 31.4% less strong than that of yam chitinase E (1.25). Taking account of the differences in glycolation degree of glycolchitin manually prepared for each study, the differences within single figures in the values of kinetic parameters such as Km and kcat might be negligible. Accordingly Pichia-produced yam chitinase proved to be similar to yam chitinase E as to kinetic behavior.
On the basis of the kinetic data, lytic activity was also expected to be high, similarly to yam chitinase E. As expected, the Pichia-produced yam chitinase showed strong lytic activity against Fusarium and Phytophthora (Fig. and Table ). The number of protoplasts released from F. oxysporum mycelia by the action of 6.0 μm Pichia-produced yam chitinase at pH 8.0 at 30 °C for 8 h of incubation was 167 × 105, while it was 104 × 105 in case of yam chitinase E under the same experimental conditions, as obtained by recalculation of the data obtained by Arakane et al.Citation15) from 6 to 8 h (Table ). Thus, it appears that the Pichia-produced yam chitinase was better than yam chitinase E in terms of protoplast formation. Taking into account of the different homogeneities of pathogenic mycelia as substrates for each study, however, such a difference in the lytic activity might be negligible. In conclusion, the Pichia-produced yam chitinase might be useful as a bio-control agent, as well as yam chitinase E.
It should be noted for further study that P. pastoris X-33 might have a capacity to modify through cyclization. The N-terminal of the recombinant protein produced by itself like the original plants such as yam, because the N-terminus was found to be blocked.
Supplemental material
The supplemental material for this paper is available at http://dx.doi.org/10.1080/09168451.2014.885825.
Supplementary Fig.1
Download MS Power Point (310 KB)Supplementary Fig.1 caption
Download MS Word (19 KB)Acknowledgment
We are thankful to Mr Minoru Iwase for his generous help in constructing the cDNA of chitinase by deletion of the intron.
Notes
Abbreviations: Endo H, Endoglycosidase H; PR protein, pathogenesis-related protein; P. pastoris, Pichia pastoris; VTS, vacuolar targeting signal; PCR, polymerase chain reaction; PAGE, polyacrylamide gel electrophoresis; SDS, sodium dodecyl sulfate.
References
- Park JK, Morita K, Fukumoto I, Yamasaki Y, Nakagawa T, Kawamukai M, Matsuda H. Biosci. Biotechnol. Biochem. 1997;61:684–689.10.1271/bbb.61.684
- Patil RS, Ghormade V, Deshpande MV. Enzyme Microb. Technol. 2000;26:473–483.10.1016/S0141-0229(00)00134-4
- Kramer KJ, Koga D. Insect Biochem. 1986;16:851–877.10.1016/0020-1790(86)90059-4
- Fan Y, Zhang Y, Yang X, Pei X, Guo S, Pei Y. Protein Expression and Purif. 2007;56:93–99.10.1016/j.pep.2007.06.012
- Mohammadi M, Roohparvar R, Torabi M. Mycopathologia. 2002;154:119–126.10.1023/A:1016039517933
- Krishnaveni S, Liang GH, Muthukrishnan S, Manickam A. Plant Sci. 1999;144:1–7.10.1016/S0168-9452(99)00050-3
- Taira T, Toma N, Ishihara M. Biosci. Biotechnol. Biochem. 2005;69:189–196.10.1271/bbb.69.189
- Vannini A, Caruso C, Leonardi L, Rugini E, Chiarot E, Caporale C, Buonocore V. Physiol. Mol. Plant Pathol. 1999;55:29–35.
- Pirttila AM, Laukkanen H, Hohtola A. Planta. 2002;214:848–852.10.1007/s00425-001-0709-x
- Santos P, Fortunato A, Ribeiro A, Pawlowski K. Plant Biotechnol. 2008;25:299–307.10.5511/plantbiotechnology.25.299
- Kuo CJ, Liao YC, Yang JH, Huang LC, Chang CT, Sung HY. J. Agric. Food Chem. 2008;56:11507–11514.10.1021/jf8017589
- Dahiya N, Tewari R, Hoondal GS. Appl. Microbiol. Biotechnol. 2006;71:773–782.10.1007/s00253-005-0183-7
- Yano S, Honda A, Rattanakit N, Noda Y, Wakayama M, Plikomol A, Tachiki T. Biosci. Biotechnol. Biochem. 2008;72:1853–1859.10.1271/bbb.80110
- Takeo S, Hisamori D, Matsuda S, Vinetz J, Sattabongkot J, Tsuboi T. Parasitol. Int. 2009;58:243–248.10.1016/j.parint.2009.05.002
- Arakane Y, Hoshika H, Kawashima N, Fujiya-Tsujimoto C, Sasaki Y, Koga D. Biosci. Biotechnol. Biochem. 2000;64:723–730.10.1271/bbb.64.723
- Karasuda S, Tanaka S, Kajihara H, Yamamoto Y, Koga D. Biosci. Biotechnol. Biochem. 2003;67:221–224.10.1271/bbb.67.221
- Mitsunaga T, Iwase M, Ubhayasekera W, Mowbray SL, Koga D. Biosci. Biotechnol. Biochem. 2004;68:1508–1517.10.1271/bbb.68.1508
- Cereghino JL, Cregg JM. FEMS Microbiol. Rev. 2000;24:45–66.10.1111/fmr.2000.24.issue-1
- Saito A, Sako Y, Usui M, Azakami H, Kato A. Biosci. Biotechnol. Biochem. 2003;67:2334–2343.10.1271/bbb.67.2334
- Lowry OH, Rosenburg NJ, Farr AL, Randall RJ. J. Biol. Chem. 1951;193:265–275.
- Imoto T, Yagishita K. Agric. Biol. Chem. 1971;35:1154–1156.10.1271/bbb1961.35.1154
- Britton HTS, Robinson RA. J. Chem. Soc. 1931;1931:1456–1462.10.1039/jr9310001456
- Lineweaver H, Burk D. J. Am. Chem. Soc. 1934;56:658–666.10.1021/ja01318a036
- Laemmli UK. Nature. 1970;227:680–685.10.1038/227680a0
- Davis BJ. Ann. N.Y. Acad. Sci. 1964;121:404–427.
- Koga D, Hirata T, Sueshige N, Tanaka S, Ide A. Biosci. Biotechnol. Biochem. 1992;56:280–285.10.1271/bbb.56.280