Abstract
β-defensins are a group of cysteine-rich cationic antimicrobial peptides that play antibacterial and antiviral roles in immune systems of vertebrates. Here, we report the cloning and identification of a β-defensin 3 cDNA sequence from the common carp (Cyprinus carpio L.). Sequence alignment and phylogenetic analysis indicated that this β-defensin 3 belonged to the BD-2 group of fish. Real-time PCR showed that the β-defensin 3 mRNA was expressed in all the tissues of normal common carp that we examined and was highly expressed in the spleen and gills. When challenged with Vibrio anguillarum, the expression level of common carp β-defensin 3 mRNA was quickly upregulated in various tissues. Our results indicate that the β-defensin 3 showed markedly high constitutive expression in the gills, and significantly upregulated expression in the hindgut of the common carp after infection, suggesting it plays an important role in the innate and mucosal immunity of common carp.
Defensins are a group of small, cationic, amphipathic, cysteine-rich antimicrobial peptides (AMPs) that have important effects in the host innate immune systems of various organisms against bacteria, fungi, and certain enveloped viruses.Citation1–4) Defensins in animals contain six cysteines in the mature peptide region that form three intracellular disulphide bonds and a β-sheet structure. Vertebrate defensins can be classified into α-, β-, and θ-defensins based on their cysteine disulphide bonding.Citation3,5,6) The six cysteines in β-defensins are linked in the pattern C1-C5, C2-C4, and C3-C6, which is different from the pattern for α-defensins and θ-defensins.Citation3) β-Defensins have been found in various vertebrates, including bovine,Citation7) human,Citation8) mouse,Citation9) bird,Citation10) reptile,Citation11) and fish,Citation12) while α-defensins and θ-defensins are known only in mammals to date,Citation3,13) suggesting that the defensin family has expanded throughout evolution and that all the subfamilies may have arisen from an ancestral β-defensin gene by duplication and diversification.Citation14)
In fish, β-defensins have recently been identified in many species, including zebrafish (Danio rerio) zfDB-1, -2, and -3; fugu (Takifugu rubripes) fuDB-1; tetraodron (Tetraodon nigroviridis) tnDB-1 and -2;Citation12) rainbow trout (Oncorhynchus mykiss) omDB-1 to -4;Citation15,16) olive flounder (Paralichthys olivaceus) fBDI-1 to -5;Citation17) gilthead seabream (Sparus aurata) saBD;Citation18) orange-spotted grouper (Epinephelus coioides) EcDefensin;Citation19) mandarin fish (Siniperca chuatsi) ScBD;Citation20) and the common carp (Cyprinus carpio L.) BD1 and BD2.Citation21) Based on homology comparison and phylogenetic analysis, the multiple defensin-like genes discovered in fish have been classified into two subgroups, the BD-1 group, and another group containing BD-2 and BD-3.Citation12,15)
In healthy fish, some of these β-defensins are expressed in a wide range of mucosal and systemic tissues, including the gills, gonad, gut, peritoneal leucocytes, kidney, liver, muscle, skin, and spleen.Citation12,16,18–20) In contrast, some β-defensins in other species have been detected only in one or two tissues.Citation12,15,21) After challenge with bacteria and with bacterial DNA, the gene expression of β-defensin was upregulated in some of these tissues,Citation16–18) suggesting the important role of this gene in the immune responses of fish. Moreover, the recombinant β-defensin peptides showed strong antimicrobial activity against gram-negative bacteria E. coli, V. anguillarum, and A. hydrophila,Citation17–20) gram-positive bacteria B. subtilis and S. saureus,Citation18,20) and certain viruses.Citation15,19)
The immune system of vertebrates is composed of two subdivisions, the innate immune system and the adaptive immune system, and innate immunity provides the first line of defense against invading pathogens. In fish, the adaptive immune system is less well developed than in higher vertebrates, and hence innate system is vitally important. Release of various AMPs is a major mechanism of the innate immune system. This acts very quickly and exerts its effects by disrupting the integrity of bacterial cell membranes or affecting intracellular targets. Thus, research on AMPs in common carp is helpful in understanding the mechanism of the innate immune system and in protecting fish from various infectious diseases. To date, several kinds of AMPs have been reported for the common carp, including two hydrophobic AMPs isolated from the skin mucus,Citation22) a hepcidin cDNA,Citation23) and two β-defensin genes (BD1 and BD2).Citation21) In the present study, we cloned and identified another β-defensin (β-defensin 3) cDNA sequence in the common carp and analyzed its expression pattern in healthy fish and fish challenged with V. anguillarum. Our results indicate that common carp β-defensin 3 might play a role in the innate immunity and is involved in the mucosal immune responses to invading pathogens.
Materials and methods
Fish rearing
Common carp (C. carpio L.) were purchased from the Fresh Water Fishery Research Institute of Shandong Province, China. These fish, weighing about 75 g each, were reared in a tank at 25 °C and fed with fish feed daily. After rearing for more than 1 week, the fish were used in experiments. The protocol for this study was approved by the Ethics Committee on Animal Experiments of the Medical School of Shandong University (Permit no. ECAESDUSM 20123009). All operations were performed under anesthesia, and all efforts were made to minimize the suffering of the fish.
Bacterial challenge
V. anguillarum (CCTCCM 204067), purchased from the China Center for Type Culture Collection, was incubated at 28 °C overnight in Luria-Bertani medium containing 3% NaCl with shaking. In the injection challenge, fish were injected intraperitoneally with formaldehyde-inactivated V. anguillarum at 5 × 107 CFU per fish. After challenge, all the fish were placed in a rectangular tank of fresh water. At 3, 12, and 24 h after challenge, three fish from each group were euthanized, and the liver, spleen, head kidney, gills, skin, foregut, and hindgut were sampled for total RNA extraction, while three unchallenged fish served as control.
RNA extraction and cDNA preparation
Tissue samples of fish were collected and powdered with a mortar in liquid nitrogen, and then total RNAs were extracted by means of RNAiso plus reagent (Takara, Dalian, China). The concentration of total RNA was measured by spectrophotometry, and quality was assessed by 1% agarose electrophoresis. Reverse transcription of mRNA and synthesis of first-strand cDNA was done by M-MLV Reverse Transcriptase (Fermentas, Burlington, Canada).
RT-PCR
Amplification of the β-defensin 3 cDNA fragments in the common carp was done by RT-PCR. Primers (shown in Table ) were designed based on the cDNA sequence of zebrafish β-defensin 3 (GenBank accession no. NM_001081555). The liver cDNA of the common carp was used as template. PCR was performed with the following settings: denaturation at 94 °C for 5 min, followed by 30 cycles of 94 °C for 30 s, annealing for 45 s, and 72 °C for 1 min, with a final extension step of 72 °C for 10 min. The annealing temperature of the primers is shown in Table too. PCR products were detected on 1% agarose gel, and the anticipated fragments were purified from the gel. The fragments were ligated into the pMD18-T vector (Takara, Dalian, China) and transformed into competent E. coli DH-5α cells, and then recombinants were identified and sequenced (Invitrogen, Shanghai, China).
Table 1. Primers used to amplify common carp β-defensin 3.
Real-time PCR
Real-time PCR analysis of β-defensin 3 gene expression was done with SYBR Green RealMasterMix (Tiangen, Beijing, China) with an iQ5 Real-time PCR instrument (Bio-Rad, Hercules, CA). The amplification scheme was: incubation for 1 min at 94 °C, followed by 40 cycles of 20 s at 94 °C, 20 s for annealing and 50 s at 70 °C. For each mRNA, gene expression was corrected by 40S ribosomal protein S11 in each sample. Relative expression of the common carp β-defensin 3 mRNA was determined by the 2(−ΔΔCt) method. In all cases, each PCR was performed with triplicate samples. The primers used and their annealing temperatures are shown in Table .
Sequence analysis, alignment, and phylogenetic analysis
The signal peptide and prodomain cleavage sites of the common carp β-defensin 3 peptide were predicted using the ProP 1.0 server (http://www.cbs.dtu.dk/services/ProP). Amino acid sequence alignment of β-defensin 3 was done with Clustal W, and a phylogenetic tree was generated by the neighbor-joining method in MEGA 5.2. The GenBank accession numbers for these sequences are shown in Table .
Table 2. GenBank accession numbers for β-defensin and big defensin sequences.
Statistical analysis
Differences in relative gene expression between the V. anguillarum-challenged group and the control group were tested by two-way analysis of variance (ANOVA) in Graphpad Prism 5, and were considered significant at p < 0.05.
Results and discussion
Common carp β-defensin 3 cDNA and deduced amino acid sequence
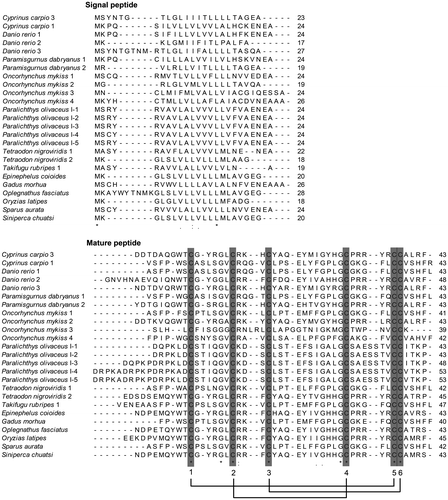
In the present study, we cloned and identified a novel β-defensin cDNA sequence from the head kidney of the common carp (C. carpio L.) by the RT-PCR method. The complete CDS of the β-defensin cDNA (GenBank accession No. KF321773) was 201 bp long encoding a peptide of 66 amino acids. The deduced amino acid sequence of common carp β-defensin 3 included two domains: a signal peptide of 23 amino acids and a mature peptide of 43 amino acids with six cysteins at the conserved sites (Fig. ). According to the results of BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and sequence alignment (data not shown), this novel β-defensin in the common carp was β-defensin 3. Similarly to other reported β-defensins, the common carp β-defensin 3 mature peptide possessed characteristic features of small size and a net cationic charge, the molecular weight of which was 5125 Da and the isoelectric point 8.28.
Sequence alignment and phylogenetic analysis of β-defensin peptide
Sequence alignment of the common carp β-defensin 3 peptide with other reported β-defensins in fish species was done at the amino acid level (Fig. ). The GenBank accession numbers of these sequences are shown in Table . The deduced amino acid sequence from the common carp β-defensin 3 cDNA was highly similar to the other fish β-defensins, and shared six cysteine residues at the identical conserved position of the mature peptide, which formed three intramolecular disulphide bonds through the linking pattern C1-C5, C2-C4, and C3-C6 (Fig. ). The percentage identities of the common carp β-defensin 3 with the other β-defensins amino acid sequences are shown in Table .
Fig. 2. Amino acid sequence alignment of β-defensin 3.
Notes: Alignment was done by Clustal W. Identical amino acid sites of these sequences are denoted by (*). Three intramolecular disulphide bonds are denoted by single lines. The GenBank accession numbers for these sequences are shown in Table .
To determine the phylogenetic relations of β-defensins in various species, a phylogenetic tree was constructed with the amino acid sequences of the β-defensins. In the tree, except for rainbow trout DB-3, all the bony fish β-defensins appeared in a branch separated from the branches of non-fish species, including mammalians, birds, and reptilians.
Fish defensins can be classified into two sub groups, the BD-1 group (containing a DB-4 of rainbow trout) and the BD-2 group, which contained zebrafish DB-3 and the common carp β-defensin 3 identified in this study (Fig. ). Additionally, several big defensins, as a homologous family of β-defensins in invertebrate, appeared in a branch position with the fish BD-2 group, indicating higher homology with BD-2 in fish.
Fig. 3. Phylogenetic tree of β-defensin 3 amino acid sequences.
Notes: This tree was generated by neighbor-joining method in MEGA 5.2. Numbers above the nodes represent boot-strap percentages of 1000 replicates. The GenBank accession numbers of these sequences are shown in Table . Common carp (C. carpio) β-defensin 3 is denoted by (◆).
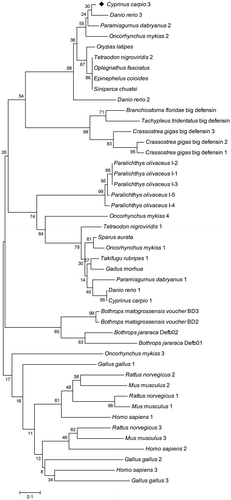
The BD-2 group in fish possesses two characteristics: five amino acid residues between Cys 1 and Cys 2, and the PRRYR motif between Cys 4 and Cys 5 (which in the O. mykiss omDB-2 molecule is PRRLR). On the other hand, there are six amino acid residues between Cys 1 and Cys 2 in the BD-1 cluster, and the motif between Cys 4 and Cys 5 is GKGLF/Q (which in the O. mykiss omDB-4 is AKGFV and in G. morhua β-defensin is GKEFQ). Absolutely different from these β-defensin sequences in fish, the motif in P. olivaceus fBDI-1 to -5 is SAESSTV, and in O. mykiss omDB-3 is TEPNV. Accordingly, in the phylogenetic tree, five P. olivaceus fBDIs form a single branch in the BD-1 cluster, and O. mykiss omDB-3 is separate from all the other fish β-defensins.
Expression distribution of the β-defensin 3 gene in the normal common carp
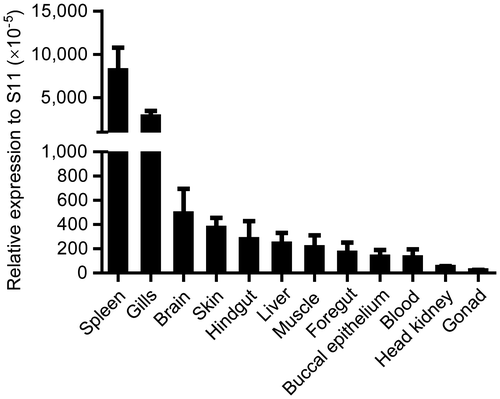
Real-time PCR was performed to analyze the expression distribution of the β-defensin 3 gene in various tissues of the normal common carp. The results indicated that the β-defensin 3 mRNA was highly expressed in the spleen and gills, and expressed in all the other examined tissues, including the brain, skin, hindgut, liver, muscle, foregut, buccal epithelium, blood, head kidney, and gonad (Fig. ), similarly to the results for zebrafish,Citation12) rainbow trout,Citation16) gilthead seabream,Citation18) orange-spotted grouper,Citation19) and mandarin fish.Citation20) Among these tissues, the common carp β-defensin 3 mRNA was mainly expressed in the spleen, which coincided with omBD-4 in the rainbow trout and ScBD in the mandarin fish. Additionally, there was high expression of β-defensin in the gills of the common carp (β-defensin 3), rainbow trout (omDB-3 and -4), orange-spotted grouper, and gilthead seabream. However, some other β-defensins, for example, zebrafish zfDB-2Citation12), rainbow trout omBD-1, and -2,Citation15,16) and common carp BD1 and BD2,Citation21) were detected only in one or two tissues. In the common carp, expression of BD1 was detected only in the skin, and BD2 only in the liver. The wide expression of β-defensin 3 in the common carp indicates that it might play an important role in the innate immune response of the common carp against bacterial infection. Furthermore, the results indicated that besides a systemic lymphoid tissue, the spleen, β-defensin 3 was also highly expressed in a mucosal lymphoid tissue, the gills, suggesting a role of β-defensin 3 in the mucosal immune system of the common carp.
Fig. 4. Tissue expression of common carp β-defensin 3 in normal common carp.
Notes: β-defensin 3 transcripts in the spleen, gills, brain, skin, hindgut, liver, muscle, foregut, buccal epithelium, blood, head kidney, and gonad of the common carp were detected by real-time PCR. 40S ribosomal protein S11 in each tissue was amplified as internal control.
β-Defensin 3 expression pattern in the common carp after bacterial challenge
In order to investigate the immune response of common carp β-defensin 3, we analyzed the changes in β-defensin 3 expression levels in the common carp under bacterial challenge. The expression level of β-defensin 3 mRNA in the common carp after i.p. injection with V. anguillarum was upregulated quickly in all examined tisssues (p < 0.05, p < 0.01 or p < 0.001), except for the liver. However, in the liver of the orange-spotted grouper, EcDefensin transcripts were significantly upregulated after injection with LPS,Citation19) differently from our results for the liver of the common carp. The liver of fish can be considered an immune organ, which secretes various immune molecules into the blood (systemic immunity) or the bile (mucosal immunity). The different expression patterns of β-defensin in the liver of different fish species and the function of the liver in the immune system call for further study.
As two important systemic immune organs, the spleen and the head kidney play key roles in the immune responses of fish against invading pathogens. Expression of β-defensin 3 in the spleen and the head kidney of the common carp was induced markedly in the present study, and the highest induction level occurred at 12 h at 12.8-fold and at 3 h at 1.7-fold after infection (Fig. ). Similarly to these results, the orange-spotted grouper EcDefensin transcripts were significantly upregulated at different post-injection times in the spleen after injection of LPS,Citation19) and induction of olive flounder fBDI mRNA was also observed in the head kidney after immune stimulation with E. tarda.Citation17) The quick upregulation in the systemic immune organs of the common carp suggests that β-defensin is involved in the innate immune response of the common carp against bacterial pathogens.
Fig. 5. Relative expression of β-defensin 3 in the common carp after i.p. injection with V. anguillarum.
Notes: Relative expression of β-defensin 3 in the liver, spleen, head kidney, gills, skin, foregut, and hindgut of the common carp at different time points is shown; n = 3. These results were normalized by means of 40S ribosomal protein S11. Data are presented as fold increase of the V. anguillarum-challenged group in relation to the uninfected group, and are shown as mean ± SD. p < 0.05, **p < 0.01 and ***p < 0.001 vs. uninfected fish.
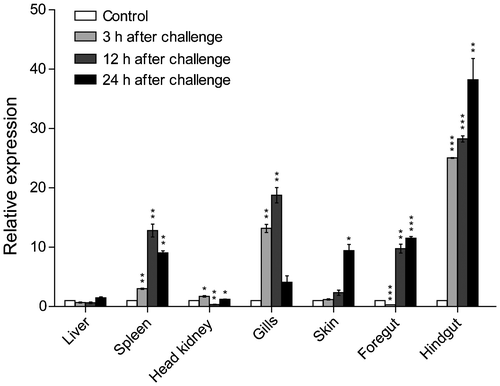
Furthermore, in our study, the expression levels of β-defensin 3 mRNA in the gills, skin, foregut, and hindgut of the common carp were upregulated significantly, and the highest induction levels occurred at 12 and 24 h after infection. Similarly, in rainbow trout, four defensin genes were induced in the gut and two other mucosal sites examined (the skin and gills) after challenge with Y. ruckeri for 48 h.Citation16) In the common carp, the expression levels of BD2 in the gills and both BD1 and BD2 in the skin significantly increased under a β-glucan feeding regimen.Citation21) The skin, gills, foregut, and hindgut are important mucosal immunity sites in fish, and they serve as the first barrier against infection.Citation24) Among these tissues, the expression of β-defensin 3 mRNA was drastically elevated, by 38.2-fold, in the hindgut. This was much higher than that in the skin (9.4-fold), the gills (18.8-fold), or the foregut (11.5-fold). These results indicate that β-defensin might play a role in the mucosal immune system, especially in the hindgut of the common carp.
Injection of V. anguillarum intraperitoneally induced the expression of β-defensin 3 in various tissues of the common carp, indicating that bacteria may not directly induce its expression. What factors are involved in the upregulation of AMPs? Cuesta et al. reported that TLR9 was involved in saBD gene activation in the gilthead seabream,Citation18) and the results of Zhao et al. revealed that NF-κB and Sp1 are responsible for immune modulation of medaka β-defensin.Citation25) Hence, we speculate that some innate immune signaling pathways and induced cytokines are involved in the regulation of AMP expression in fish after challenge.
Conclusion
To summarize, in this study, we cloned and identified a β-defensin 3 cDNA sequence from the common carp, and analyzed the phylogenetic relation and expression pattern of common carp β-defensin 3 under challenge by bacterium. The results indicate that β-defensin 3 might play an important role in the innate immune response, especially in the hindgut and gills mucosal immune responses of the common carp to bacterial pathogens. However, the antimicrobial activity of purified, synthetic, or recombinant common carp β-defensin 3 against different bacteria and viruses call for further research. All this research should be of importance in uncovering the innate immune mechanism of fish, and helpful in protecting the common carp against many bacterial diseases in carp farming.
Author contribution
Conceived and designed the experiments: Liguo An and Guiwen Yang. Performed the experiments: Hua Li, Hongyan Guo, Shijuan Shan, and Chenchen Qi. Analyzed the data: Hua Li, Hongyan Guo, Liguo An, and Guiwen Yang. Wrote the paper: Hua Li and Guiwen Yang.
Acknowledgments
This work was supported by the University Independent Innovation Project of Jinan, Shandong, China [grant numbers 200715072 and 200906020].
Notes
Abbreviations: AMPs, antimicrobial peptides; BD, β-defensin; CFU, colony forming units; RT-PCR, reverse transcription-polymerase chain reaction; CDS, coding sequence.
References
- Bulet P, Stocklin R. Insect antimicrobial peptides: structures, properties and gene regulation. Protein Pept. Lett. 2005;12:3–11.10.2174/0929866053406011
- Castro MS, Fontes W. Plant defense and antimicrobial peptides. Protein Pept. Lett. 2005;12:13–18.
- Ganz T. Defensins: antimicrobial peptides of innate immunity. Nat. Rev. Immunol. 2003;3:710–720.10.1038/nri1180
- Zhang L, Yu W, He T, Yu J, Caffrey RE, Dalmasso EA, Fu S, Pham T, Mei J, Ho JJ, Zhang W, Lopez P, Ho DD. Contribution of human alpha-defensin 1, 2, and 3 to the anti-HIV-1 activity of CD8 antiviral factor. Science. 2002;298:995–1000.10.1126/science.1076185
- Lynn DJ, Higgs R, Gaines S, Tierney J, James T, Lloyd AT, Fares MA, Mulcahy G, O’Farrelly C. Bioinformatic discovery and initial characterisation of nine novel antimicrobial peptide genes in the chicken. Immunogenetics. 2004;56:170–177.10.1007/s00251-004-0675-0
- Xiao Y, Hughes A, Ando J, Matsuda Y, Cheng J-F, Skinner-Noble D, Zhang G. A genome-wide screen identifies a single β-defensin gene cluster in the chicken: implications for the origin and evolution of mammalian defensins. BMC Genomics. 2004;5:56–66.10.1186/1471-2164-5-56
- Diamond G, Zasloff M, Eck H, Brasseur M, Maloy WL, Bevins CL. Tracheal antimicrobial peptide, a cysteine-rich peptide from mammalian tracheal mucosa: peptide isolation and cloning of a cDNA. Proc. Natl. Acad. Sci. USA. 1991;88:3952–3956.10.1073/pnas.88.9.3952
- Bensch KW, Raida M, Magert HJ, Schulz-Knappe P, Forssmann WG. hBD-1: a novel β-defensin from human plasma. FEBS Lett. 1995;368:331–335.10.1016/0014-5793(95)00687-5
- Huttner KM, Kozak CA, Bevins CL. The mouse genome encodes a single homolog of the antimicrobial peptide human β-defensin 1. FEBS Lett. 1997;413:45–49.10.1016/S0014-5793(97)00875-2
- Evans EW, Beach GG, Wunderlich J, Harmon BG. Isolation of antimicrobial peptides from avian heterophils. J. Leukocyte Biol. 1994;56:661–665.
- Rádis-Baptista G, Oguiura N, Hayashi MAF, Camargo ME, Grego KF, Oliveira EB, Yamane T. Nucleotide sequence of crotamine isoform precursors from a single South American rattlesnake (Crotalus durissus terrificus). Toxicon. 1999;37:973–984.10.1016/S0041-0101(98)00226-8
- Zou J, Mercier C, Koussounadis A, Secombes C. Discovery of multiple beta-defensin like homologues in teleost fish. Mol. Immunol. 2007;44:638–647.10.1016/j.molimm.2006.01.012
- Yang D, Biragyn A, Hoover DM, Lubkowski J, Oppenheim JJ. Multiple roles of antimicrobial defensins, cathelicidins, and eosinophil-derived neurotoxin in host defense. Annu. Rev. Immunol. 2004;22:181–215.10.1146/annurev.immunol.22.012703.104603
- Semple CAM, Rolfe M, Dorin JR. Duplication and selection in the evolution of primate b-defensin genes. Genome Biol. 2003;4:R31.10.1186/gb-2003-4-5-r31
- Falco A, Chico V, Marroqui L, Perez L, Coll JM, Estepa A. Expression and antiviral activity of a beta-defensin-like peptide identified in the rainbow trout (Oncorhynchus mykiss) EST sequences. Mol. Immunol. 2008;45:757–765.10.1016/j.molimm.2007.06.358
- Casadei E, Wang T, Zou J. Characterization of three novel β-defensin antimicrobial peptides in rainbow trout (Oncorhynchus mykiss). Mol. Immunol. 2009;46:3358–3366.10.1016/j.molimm.2009.07.018
- Nam BH, Moon JY, Kim YO, Kong HJ, Kim WJ, Lee SJ, Kim KK. Multiple β-defensin isoforms identified in early developmental stages of the teleost Paralichthys olivaceus. Fish Shellfish Immunol. 2010;28:267–274.10.1016/j.fsi.2009.11.004
- Cuesta A, Meseguer J, Esteban MA. Molecular and functional characterization of the gilthead seabream β-defensin demonstrate its chemotactic and antimicrobial activity. Mol. Immunol. 2011;48:1432–1438.10.1016/j.molimm.2011.03.022
- Guo M, Wei J, Huang X, Huang Y, Qin Q. Antiviral effects of β-defensin derived from orange-spotted grouper (Epinephelus coioides). Fish Shellfish Immunol. 2012;32:828–838.10.1016/j.fsi.2012.02.005
- Wang G, Li J, Zou P, Xie H, Huang B, Nie P, Chang M. Expression pattern, promoter activity and bactericidal property of β-defensin from the mandarin fish Siniperca chuatsi. Fish Shellfish Immunol. 2012;33:522–531.10.1016/j.fsi.2012.06.003
- van der Marel M, Adamek M, Gonzalez SF, Frost P, Rombout JH, Wiegertjes GF, Savelkoul HF, Steinhagen D. Molecular cloning and expression of two β-defensin and two mucin genes in common carp (Cyprinus carpio L.) and their up-regulation after β-glucan feeding. Fish Shellfish Immunol. 2012;32:494–501.10.1016/j.fsi.2011.12.008
- Lemaitre C, Orange N, Saglio P, Saint N, Gagnon J, Molle G. Characterization and ion channel activities of novel antibacterial proteins from the skin mucosa of carp (Cyprinus carpio). Eur J Biochem. 1996;240:143–149.10.1111/(ISSN)1432-1033
- Li H, Zhang F, Guo H, Zhu Y, Yuan J, Yang G, An L. Molecular characterization of hepcidin gene in common carp (Cyprinus carpio L.) and its expression pattern responding to bacterial challenge. Fish Shellfish Immunol. 2013;35:1030–1038.10.1016/j.fsi.2013.07.001
- Magnadottir B. Innate immunity of fish (overview). Fish Shellfish Immunol. 2006;20:137–151.10.1016/j.fsi.2004.09.006
- Zhao JG, Zhou L, Jin JY, Zhao Z, Lan J, Zhang YB, Zhang QY, Gui JF. Antimicrobial activity-specific to Gram-negative bacteria and immune modulation-mediated NF-κB and Sp1 of a medaka β-defensin. Dev. Comp. Immunol. 2009;33:624–637.10.1016/j.dci.2008.11.006

