Abstract
We identified and analyzed a DNA region that is required for the stable maintenance of plasmids in the genus Sphingomonas. This DNA fragment, a 244 bp, is localized in the upstream region of the repA gene of low-copy-number small plasmid pYAN-1 (4896 bp) of Sphingobium yanoikuyae. It has four inverted repeats and one direct repeat for possible secondary structures. We were able to stabilize not only another unstable plasmid, pYAN-2, in the genus Sphingomonas, but also the unstable plasmid pSC101 without par locus in Escherichia coli. The copy-number levels between the unstable plasmid and the parental plasmid were similar, and these results suggest that the stabilization of unstable plasmids by this DNA region of pYAN-1 was not due to an increase in plasmid copy number. We concluded that the stabilization of the plasmid was due to a plasmid partition mechanism encoded by a DNA fragment of pYAN-1.
Graphical Abstract
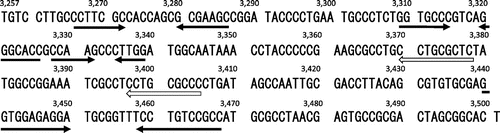
Nucleotide sequence of a non-coding DNA region of pYAN-1 that is required for the stable maintenance of plasmids in Sphingomonas and Escherichia coli.
Key words:
Low-copy-number bacterial plasmids encode partition (Par) systems that ensure active segregation of plasmid copies prior to cell division. Most Par systems consist of three elements: an NTPase (generally ATPase), a DNA-binding protein, and a centromere. The DNA-binding protein, generically termed ParB, interacts with the centromere, parS, to create a partition complex. The NTPase, ParA, is recruited by the partition complexes to move sister plasmids toward opposite cell poles. There are two main types of Par system defined according to the nature of the ATPase.Citation1,2) Type II, currently the best understood, has an actin-like ATPase that pushes sister plasmids to the poles by forming filaments.Citation3,4) Type I, the most common, has a P-loop ATPaseCitation5) that oscillates over the nucleoidCitation6,7) and, at least in vitro, polymerizes.Citation8,9) Type I ParA and ParB proteins are diverse in sequence,Citation10) but their genetic organization falls mostly into two subtypes: Ia (e.g. plasmids F and P1) and Ib (e.g. plasmids pTAR, TP228, and pB171-Par2).Citation11) Ia ParA proteins are large, with an N-terminal extension that binds the promoter regulating the parAB operon, while Ib ParA proteins are small and have no promoter-binding domain. Ia ParB proteins bind the centromere via a helix-turn-helix domain. Ib ParB proteins are small and bind via a ribbon-helix-helix domain to a centromere that harbors the parAB promoter.Citation12) More recently, type III and IV partition systems have been identified. The type III systems drive partition with tubulin-like GTPases, whereas the type IV systems use a single, non-NTPase coiled-coil protein.Citation13–15) All types of plasmid maintenance, I–IV systems, are found in large and giant plasmids, but there have been no studies on the stabilization of small low-copy-number plasmids. On the other hand, several studies have found that the partition function, termed par, of pSC101 in Escherichia coliCitation16) and in pLS11,Citation17) also referred to as pPOD2000Citation18) in Bacillus subtilis, restores partition ability. These par sequences act only in cis, encode no protein, and stabilize unrelated plasmids. Partitioning of pSC101 is not dependent on the location or orientation on the plasmid of the reintroduced par fragment, but that of pLS11 is orientation dependent.
Recently, the genus Sphingomonas has received increasing attention because it includes various xenobiotic-degrading bacteria. The members of this genus degrade compounds such as polycyclic aromatic hydrocarbons, chlorinated and sulfonated aromatics, herbicides, aromatic ethers, and polyethylene glycol.Citation19) There have been several reports indicating that giant plasmids are important in the degradation of xenobiotic compounds by Sphingomonas strains.Citation20,21) In Sphingomonas aromaticivorans F199 and some other sphingomonads isolated from the same location, genes encoding pathways for degrading biphenyl, naphthalene, m-xylene, and p-cresol have been detected in megaplasmids. Moreover, strains of the genus Sphingomonas have a unique characteristic: they contain glycosphingolipids, which are ubiquitous in eukaryotic cell membranes.Citation22) When Sphingomonas sp. A1 assimilates a macromolecule (alginate), a mouth-like pit (0.02–0.1 μm) forms on the cell surface through reorganization and/or the fluidity of the pleats, causing extracellular alginate to be concentrated in the pit.Citation23) This pit-dependent system of importing macromolecules was reported for the first time in a prokaryote, but appears to be the origin of endocytosis and phagocytosis in eukaryotes. Genetic manipulation of the genus Sphingomonas is necessary to improve the capacity of these bacteria to degrade xenobiotic compounds and to determine the unique mechanisms involved in degradation, but no plasmid suitable for genetic manipulation of Sphingomonas has yet been developed. Some vector systems in sphingomonads with a broad-host-range plasmid or a cryptic giant plasmid have been achieved by the conjugation method.Citation19,24) We have reported the isolation and characterization of three small plasmids, pAMI-1 from Sphingobium amiense JCM11777Citation25) and pYAN-1 and pYAN-2 from Sphingobium yanoikuyae JCM7371,Citation26) along with the development of a transformation system.
In the present study, we identified and characterized a DNA region for a plasmid maintenance system derived from pYAN-1, and this DNA region increased the stability of other unstable plasmids in cis. This is the first report on a stabilization system based on a DNA region of a low-copy-number small plasmid.
Materials and methods
Bacteria, plasmids, and media
Novosphingobium aromaticivorans F199Citation20) was cultured routinely at 30 °C in LB medium and used as host strain for the plasmids. E. coli DH5α [(80dlacZΔM15) endA1 recA1 hsdR17(r−m−) supE44 thi-1 λ− gyrA relA1 F− Δ(lacZYA-argF) U169] strains were grown at 37 °C in LB medium and were used as hosts in the construction of plasmids and in routine subcloning. Plasmids pYAN-1 and pYAN-2 were prepared from S. yanoikuyae JCM7371.Citation26) Plasmid pHSG399, carrying the chloramphenicol resistance gene, was purchased from Takara Shuzo (Kyoto, Japan).Citation27) Plasmid pMW119,Citation28) carrying the ampicillin resistance gene, was from Nippon Gene (Tokyo). E. coli transformants were selected by resistance to 20 μg/mL of chloramphenicol (Cm) or 100 μg/mL of ampicillin (Ap). Transformed Sphingomonas strains were selected based on resistance to 15 μg/mL of Cm.
Plasmid stability test
Cells harboring the plasmids were inoculated in LB supplemented with selective antibiotics, and grown at 30 °C to the stationary phase. At this point, cultures were diluted 105-fold in fresh LB without antibiotics and grown through 15–20 generations. Samples of each culture were taken at the beginning and end of growth, diluted, spread on LB agar plates without antibiotics, and grown to 100–300 colonies per plate. The phenotypes of 100 colonies from each plate were examined by transferring them with toothpicks to selection plates containing antibiotics. Cell concentrations were determined before and after cultivation. To calculate standard deviation, all experiments were repeated at least five times.
DNA manipulation
Plasmid DNA was prepared from transformants of Sphingomonas by a modification of the alkaline lysis procedure.Citation29) Approximately, 109 cells were harvested by centrifugation, washed with 1 mL of G buffer (50 mM Tris-HCl pH 8.0, 1 mM EDTA pH 8.0, and 10% vol/vol glycerol), resuspended in 100 μL of lysozyme solution (25 mM Tris-HCl pH 8.0, 1 mM EDTA pH 8.0, 100 mM NaCl, 15% wt/vol sucrose, and 0.8% wt/vol lysozyme), and incubated at 37 °C for 20 min. The cell suspensions were gently mixed with 200 μL of solution II (0.2 M NaOH and 1.0% wt/vol SDS) and incubated at room temperature until the cells were lysed. Then, the cell lysates were mixed with 150 μL of solution III (3 M potassium acetate and 11.5% vol/vol glacial acetic acid) and left on ice for 5 min. Finally, plasmid DNA was prepared by phenol/chloroform extraction and precipitation in ethanol and dissolved in TE buffer (10 mM Tris-HCl pH 8.0 and 1 mM EDTA) containing 10 μg/mL of RNase A.
Electroporation
Cells from 50 mL cultures of Sphingomonas strains (optical density at 660 nm, 0.7–0.8) were collected by centrifugation and washed twice with 10 mL of chilled 10% glycerol. The cells were resuspended in the same buffer to a final volume of 100 μL and mixed with plasmid DNA (1 μg) and placed in 0.2 cm cuvettes. Electroporation was done with a Gene Pulser (Bio-Rad, Hercules, CA) with a single pulse at 25 μF at 2.5 kV. The cells were allowed to grow in LB for 2 h, and then were spread on selection plates containing antibiotics for selection.
Results and discussion
Identification of the DNA region for plasmid maintenance
The cryptic small plasmid pYAN-1 (4896 bp) was originally isolated from S. yanoikuyae JCM7371 by Ochou et al.Citation26) We are unaware of any selection pressure that selects for the presence of this plasmid in the bacterial population, and we suppose that pYAN-1 contains within its genome a genetic element(s) that control plasmid maintenance. The previous reports described above indicate that all types of plasmid maintenance, the I–IV systems, are found near the replication region in various low-copy-number plasmids. Moreover, all the par sequences described above are found in upstream regions of the repA gene encoding the replication protein. Still, we could not find the typical Par system in the pYAN-1 genome, and we speculate that pYAN-1 is stable due to a par-like sequence adjacent to the upstream region of the repA gene.
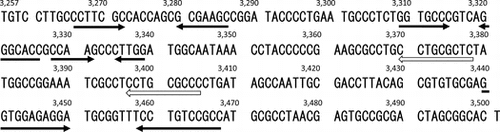
First, we determined the minimal replication region surrounding the repA gene of pYAN-1. We constructed a shuttle plasmid, pYAN1-A, between E. coli and Sphingomonas, with the upstream regions surrounding the repA gene from pYAN-1 amplified by PCR with the following primers: 5-AACTGCAGGCTCGCCCTTCAAACC-3 (forward, PstI site underlined) and 5-AAGAATTCCTTCTCTTAGGGTG-3 (reverse, EcoRI site underlined) (the fragment from nt 3122 to 4470 in Fig. ). The PCR product was digested with Pst I and EcoRI and then inserted into PstI/EcoRI-digested E. coli plasmid pHSG399, which harbors a chloramphenicol (Cm) resistance gene, and the resulting plasmid was designated pYAN1-A. pYAN1-A was maintained stably in Sphingomonas cells for at least 60 generations in the absence of selection (Fig. ). To determine the minimal region necessary for plasmid maintenance, we constructed various deletion plasmids by the method used for pYAN1-A (Fig. ). All plasmids were constructed by PCR with the above described reverse primer and various forward primers (5-AACTGCAGTGTCCTTGCCCTTC-3 for pYAN1-B containing the fragment from nt 3257 to 4470, 5′-AACTGCAGCCCGAAGCGCCTGCC-3′ for pYAN1-C containing the fragment from nt 3357 to 4470, 5-AACTGCAGCGTGTGCGAGGTGG-3 for pYAN1-D containing the fragment from nt 3431 to 4470, 5-AACTGCAGTCGTTGTTCACTTTTTGTTCC-3 for pYAN1-E containing the fragment from nt 3501 to 4470, and 5-AACTGCAGAGAAGGGGCAGA-3 for pYAN1-F containing the fragment from nt 3582 to 4470, PstI sites underlined). Four plasmids (pYAN1-B, pYAN1-C, pYAN1-D, and pYAN-1E) transformed Sphingomonas cells, but plasmid pYAN1-F did not (Fig. ). These results suggest that the replication function of pYAN-1 is located within the nt 3501 to 4470 fragment. To determine the minimal region necessary for plasmid maintenance, we examined the stability of various deletion plasmids under non-selective culture conditions. Three plasmids (pYAN1-C, pYAN1-D, and pYAN-1E) were unstable, and the rates of segregation were different (Fig. ). pYAN1-B was stable, as was pYAN1-A (Fig. ). These results indicate that the mechanism of plasmid maintenance is located within the DNA region from nt 3257 to 3501 of plasmid pYAN-1.
Fig. 1. Transformation of Sphingomonas with deletion derivatives of pYAN1-A.
Note: Plasmid construction is described in the text. The transformation method is described in “Materials and methods.”
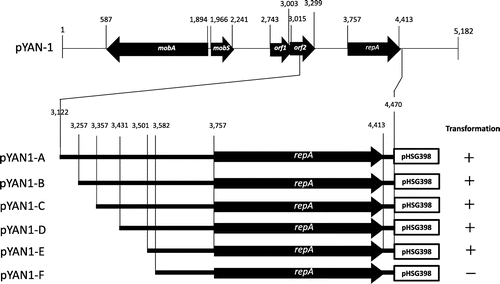
Fig. 2. Stability of various plasmids under non-selective conditions.
Note: The stability of plasmids is shown as percentages of plasmid-containing cells. The stability test is described in “Materials and methods.” Cells for the stability test were obtained from about 15, 30, 45, 60, and 75 generations of growth. To calculate standard deviation, all experiments were repeated five times. Results are averages for five experiments. Symbols: ● pYAN1-A; ○ pYAN1-B; ■ pYAN1-C; □ pYAN1-D; and ▲ pYAN1-E.
The nucleotide sequence of this region is shown in Fig. . Computational analysis by the GENETYX program (ver. 11.1.0) revealed that the fragment of pYAN-1 did not encode any significant protein sequences, and the same genes for the 167 bp fragment of pLS11Citation17) and the 375 bp fragment of pSC101.Citation30) Moreover, there was no significant sequence or secondary structure homology between the fragment of pYAN-1 and the par of pSC101 or pLS11 at the nucleotide level. As possible secondary structures, four inverted repeats and one direct repeat were detected in this fragment of pYAN-1 (Fig. ), but the functions of these structures remain unknown.
Stabilization of unstable plasmids by the DNA region of pYAN-1
We have reported that plasmid pYAN-2 was compatible with plasmid pYAN-1 in the same cells.Citation23) These results indicate that the replication systems of the two plasmids, pYAN-1 and pYAN-2, are quite different and are of different origins. First, we determined the minimal replication region surrounding the repA gene of pYAN-2. We constructed a shuttle plasmid, pYAN2-A, between E. coli and Sphingomonas, with the upstream regions surrounding the repA gene from pYAN-2 amplified by PCR by the following primers: 5-AAGGATCCGTTCATATAGTTCTT-3 (forward, BamHI site underlined) and 5-AAGAATTCTCCGCTAATCTAC-3 (reverse, EcoRI site underlined, the fragment from nt 3092 to 4367 in Fig. (A)). The PCR product was digested with BamHI and EcoRI, inserted into BamHI/EcoRI-digested E. coli plasmid pHSG399, and designated pYAN2-A. pYAN2-A was unstable in Sphingomonas cells in the absence of selection (Fig. (B)).
Fig. 4. Construction and stability of pYAN2-A and pYAN2A-244.
Note: (A) Plasmid construction is described in the text, and the stability test is described in “Materials and methods.” (B) Cells for the stability test were obtained from about 13 and 26 generations of growth. To calculate standard deviation, experiments were repeated five times. Results are averages for five experiments. Arrowheads: inverted repeats;
direct repeats. Symbols: ○ pYAN2-A; □ pYAN2A-244.
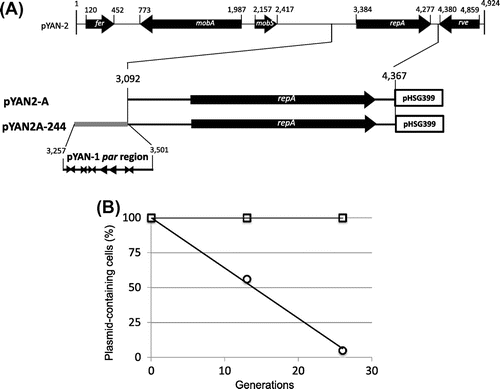
To test whether the above fragment of plasmid pYAN-1 can stabilize unstable plasmid pYAN2-A, we constructed plasmid pYAN2A-244 as follows: a fragment was amplified by PCR with the following primers: 5-AAAAGCTTTGTCCTTGCCCTTC-3 (forward, HindIII site underlined) and 5-AAAAGCTTAGTGCCGCTAGTC-3 (reverse, HindIII site underlined) for pYAN-1 (the fragment from nt 3257 to 3501 in Fig. (A)). The PCR fragment DNA was digested with HindIII and then inserted into HindIII-digested plasmid pYAN2-A (Fig. (A)). Plasmid pYAN2A-244 was stabilized for 26 generations under non-selective culture conditions (Fig. (B)).
Since it is possible that even plasmids lacking the partition mechanism can be maintained relatively stably as multi-copy-number plasmids, we measured the difference in copy number between plasmid pYAN2-A and plasmid pYAN2A-244 as the amount of plasmid DNA relative to the amount of chromosomal DNA by a previous method of ours.Citation22) The copy number of stable plasmid pYAN2A-244 was similar to that of unstable plasmid pYAN2-A, and the copy numbers were on average 1 or 2 per chromosome (data not shown). Moreover, the copy number of stable plasmid pYAN1-A was also 1 or 2 plasmids per chromosome (data not shown), and both pYAN-1 and pYAN-2 were stringently regulated for plasmid replication. These results suggest that the stabilization of plasmid pYAN2-A by this region of pYAN-1 was not due to an increase in plasmid copy number. We concluded that stabilization of the plasmid was due to a plasmid partition mechanism encoded by this fragment, and designated it par.
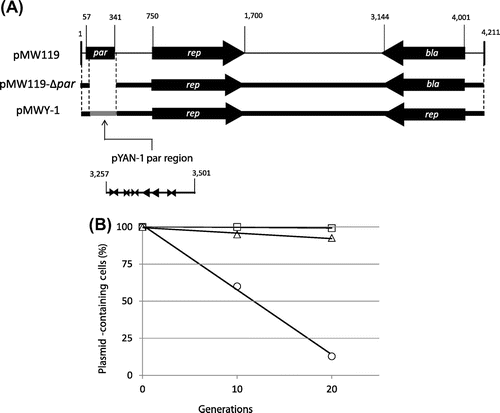
To test whether the par region of pYAN-1 can function in other bacteria, such as E. coli, we constructed an unstable E. coli plasmid with a low-copy-number plasmid. First, we constructed the unstable pMW119Citation28) plasmid (based on plasmid pSC101). The region surrounding the par gene from pMW119 was amplified by PCR with the following primers: 5-AAGATATCGCTTGCGAGG-3 (forward, EcoRV site underlined) and 5-AAGATATCTTCGGATTATCCCGTGACAGG-3 (reverse, EcoRVsite underlined, the fragment from nt 341 to 57 in Fig. (A)). The PCR fragment was digested with EcoRV and re-ligated to eliminate the par gene, and the result was designated pMW119-Δpar. pMW119-Δpar was unstable in E. coli cells for at least 20 generations in the absence of selection (Fig. (B)). To test whether the above par region of plasmid pYAN-1 can stabilize plasmid pMW119-Δpar, we constructed plasmid pMWY-1, as follows: a par region was amplified by PCR with the following primers: 5-AAGATATCTCTAAAGGGGGT-3 (forward, EcoRVsite underlined) and 5′-AAGATATCAGTGCCGCTAGTC-3′ (reverse, EcoRVsite underlined) for pYAN-1 (the fragment from nt 3257 to 3501 in Fig. (A)), and the PCR fragment DNAs were digested with EcoRV and then inserted into EcoRV-digested pMW119-Δpar plasmid (Fig. (A)). Plasmid pMWY-1, as well as parent plasmid pMW119, was perfectively stabilized for 20 generations under non-selective culture conditions (Fig. (B)). Also, the copy-number levels as between plasmids pMW119 and pMWY-1 in E. coli were similar (data not shown). These results suggest that the par of pYAN-1 can function in E. coli.
Fig. 5. Construction and stability of pMW119Δpar and pMWY-1.
Note: (A) Plasmid construction is described in the text, and the stability test is described in “Materials and methods.” (B) Cells for the stability test were obtained from about 10 and 20 generations of growth. To calculate standard deviation, experiments were repeated five times. Results are averages for five experiments. Arrowheads: inverted repeats;
direct repeats. Symbols: ○ pMW119Δpar; □ pMW119;
pMWY-1.
Meacock and Cohen have proposed that the distribution of plasmid molecules in the case of pSC101 between daughter cells at cell division is mediated by the interaction of this DNA locus with other cellular components of the partitioning system, such as the cytoplasmic membrane, and can be initiated by duplication of the par locus.Citation16) We are also interested in plasmid partition, especially as to the interaction of the par locus with the cytoplasmic membrane, and additional studies are now in progress.
In conclusion, we identified and characterized an efficient stabilization system for plasmids in the genus Sphingomonas. We believe that this will facilitate the molecular design of high-expression and high-stable cloning vectors for the expression of foreign genes in sphingomonads for industrial purposes.
Funding
This work was supported by JSPS KAKENHI [grant number 20510021].
References
- Gerdes K, Møller-Jensen J, Bugge JR. Mol. Microbiol. 2000;37:455–466.
- Godfrin-Estevenon A-M, Pasta F, Lane D. Mol. Microbiol. 2000;43:39–49.
- Campbell CS, Mullins RD. J. Cell Biol. 2007;179:1059–1066.10.1083/jcb.200708206
- Møller-Jensen J, Jensen RB, Löwe J, Gerdes K. EMBO J. 2002;21:3119–3127.10.1093/emboj/cdf320
- Ogura T, Hiraga S. Cell. 1983;32:351–360.10.1016/0092-8674(83)90454-3
- Castaing J-P, Bouet J-Y, Lane D. Mol. Microbiol. 2008;70:1000–1011.
- Hatano T, Yamaichi Y, Niki H. Mol. Microbiol. 2007;64:1198–1213.10.1111/j.1365-2958.2007.05728.x
- Barillà D, Rosenberg MF, Nobbmann U, Hayes F. EMBO J. 2005;24:1453–1464.10.1038/sj.emboj.7600619
- Bouet JY, Ah-Seng Y, Benmeradi N, Lane D. Mol. Microbiol. 2007;63:468–481.10.1111/j.1365-2958.2006.05537.x
- Bignell C, Thomas CM. J. Biotechnol. 2001;91:1–34.10.1016/S0168-1656(01)00293-0
- Gerdes K, Howard M, Szardenings F. Cell. 2010;141:927–942.10.1016/j.cell.2010.05.033
- Dubarry N, Du W, Lane D, Pasta F. Appl. Environ. Microbiol. 2010;76:1095–1102.10.1128/AEM.02123-09
- Larsen RA, et al. Genes Dev. 2007;21:1340–1352.10.1101/gad.1546107
- Simpson AE, Skurray RA, Firth N. J. Bacteriol. 2003;185:2143–2152.10.1128/JB.185.7.2143-2152.2003
- Tang M, Bideshi DK, Park H-W, Federici BA. J. Bacteriol. 2007;189:8053–8058.10.1128/JB.00908-07
- Meacock PA, Cohen SN. Cell. 1980;20:529–542.10.1016/0092-8674(80)90639-X
- Chang S, Chang SY, Gray O. J. Bacteriol. 1987;169:3952–3962.
- Gleave AP, Mountain A, Thomas CM. J. Gen. Microbiol. 1990;136:905–912.10.1099/00221287-136-5-905
- Basta T, Keck A, Klein J, Stolz A. J. Bacteriol. 2004;186:3862–3872.10.1128/JB.186.12.3862-3872.2004
- Romine MF, Stillwell LC, Wong KK, Thurston SJ, Sisk EC, Sensen T, Gaasterland T, Fredrickson JK, Saffer JD. J. Bacteriol. 1999;181:1585–1602.
- Shintani M, Urata M, Inoue K, Eto K, Habe H, Omori T, Yamane H, Nojiri H. J. Bacteriol. 2007;189:2007–2020.10.1128/JB.01486-06
- Kawasaki S, Moriguchi R, Sekiya K, Nakai T, Ono E, Kume K, Kawahara K. J. Bacteriol. 1994;176:284–290.
- Hisano T, Kimura N, Hashimoto W, Murata K. Biochem. Biophys. Res. Commun. 1996;220:979–982.10.1006/bbrc.1996.0526
- Momma K, Okamoto M, Mishima Y, Mori S, Hashimoto W, Murata K. J. Bacteriol. 2000;182:3998–4004.10.1128/JB.182.14.3998-4004.2000
- Saito M, Ikunaga Y, Ohta H, Kurusu Y. Microb. Environ. 2006;21:235–239.10.1264/jsme2.21.235
- Ochou M, Saito M, Kurusu Y. Biosci., Biotechnol., Biochem. 2008;72:1130–1133.10.1271/bbb.70813
- Takeshita S, Sato M, Toba M, Masahashi W, Hashimoto-Gotoh T. Gene. 1987;61:63–74.10.1016/0378-1119(87)90365-9
- Yamaguchi K, Masamune Y. MGG Mol. Gen. Genet. 1985;200:362–367.10.1007/BF00425718
- Bimboim HC, Doly J. Nucleic Acids Res. 1979;7:1513–1523.10.1093/nar/7.6.1513
- Miller CA, Tucker WT, Meacock PA, Gustafsson P, Cohen SN. Gene. 1983;24:309–315.10.1016/0378-1119(83)90091-4