Abstract
A codon-optimized Aspergillus niger pectin methylesterase (PME) gene was expressed in the methylotrophic yeast Canidia boidinii. The PME-producing strains showed better growth on pectin than the wild-type strains, suggesting that the PME-producing strains could efficiently utilize methyl ester moieties of pectin. On the other hand, overproduction of PME negatively affected the proliferation of C. boidinii on leaves of Arabidopsis thaliana.
Methanol has been recognized as an important constituent of the atmosphere, and one of the main sources of atmospheric methanol is plant leaves.Citation1,2) The global rate of methanol emission from plant leaves is estimated to be 100 Tg (methanol C) year−1.Citation3,4) It has been considered that methanol is produced mainly as a by-product of pectin metabolism during cell-wall synthesis.Citation5) The precursors of pectin contain numerous galacturonate methyl esters, presumably to facilitate transport through the cell wall. These methyl esters are demethylated by pectin methylesterases (PMEs) in the plant,Citation6) resulting in methanol production. Extracellular PMEs are also produced by a number of filamentous fungi.
Methylotrophic yeasts can grow on methanol as the sole source of carbon and energy. Since the methylotrophic yeast Candida boidinii was first isolated in 1969,Citation7) many methylotrophic yeasts have been isolated from plant materials.Citation8,9) Recently, we demonstrated that C. boidinii can proliferate and survive on the leaves of Arabidopsis thaliana and that the methanol assimilation pathway and peroxisome homeostasis are necessary for phyllospheric proliferation.Citation10) On the other hand, many methylotrophic yeasts have been reported to grow on pectin as a carbon source.Citation11) Previously, C. boidinii was reported to be able to grow on pectin.Citation12) During growth on pectin, pectin is hydrolyzed by PME to methanol and polygalacturonate, the basic skeleton of pectin. Each of these products is then utilized as a carbon and energy source by independent metabolic pathways. In this study, we expressed a heterologous PME-encoding gene in C. boidinii and showed that the PME-producing strains can efficiently utilize methyl ester moieties of pectin. Furthermore, we examined the effect of PME overproduction on yeast proliferation in the phyllosphere.
In the previous study, C. boidinii exhibited PME activity; however, a PME-encoding gene could not be identified in the draft genome sequence. Therefore, we decided to heterologously produce the Aspergillus niger PME (accession no. P17872), which comprises of a mature protein of 314 amino acids and an N-terminal secretory signal peptide of 17 amino acids,Citation13,14) in C. boidinii. Because highly expressed genes in C. boidinii contain an A or T nucleotide at the third position of most codons, the translation efficiency of the gene which contains a G or C nucleotide at the third position of codons is likely to be low. We observed improvement in productivity of fructosyl amino acid oxidase from Pencillium janthinelum whose original cDNA contained G or C nucleotides at the third position of most codons by codon optimization.Citation15) Therefore, the 996-bp DNA fragment corresponding to the deduced amino acid sequence of the A. niger pmeA was synthesized according to the preferred codon usage of C. boidinii by a recursive PCR technique.Citation16) The sequence was submitted to DDBJ under accession no. AB872253. The PME-encoding DNA fragment was cloned into pACT1Citation17) with SalI and PstI sites at the 5′ or 3′ ends of the PME coding sequence to yield pACTPME (for expression under the constitutive actin gene, ACT1, promoter), and into pNOTeICitation18) with Not I restriction sites to yield pAODPME (for expression under the strong methanol-inducible alcohol oxidase gene, AOD1, promoter). pACTPME and pAODPME were linearized with EcoRV, and each was introduced into C. boidinii TK62 ura3 using the previously described modified lithium acetate method.Citation19) The resulting strains, PMEact and PMEaod, were obtained, respectively. Introduction of the expression vector was confirmed by the Southern blot analysis with EcoRI-digested genomic DNA of strain PMEact and PstI-digested genomic DNA of strain PMEaod, and a 1.0-kb fragment of PME-encoding DNA fragment as the probe (data not shown).
The strains, PMEact and PMEaod were grown to mid-exponential phase in SD medium (0.67% yeast nitrogen base without amino acids and 2% glucose). Then, cells were transferred to the SM medium (0.67% yeast nitrogen base without amino acids and 1% methanol) and incubated for 16 h. Subsequently, cells were harvested by centrifugation at 12,000 × g for 10 min at 4 °C. Supernatants were concentrated using a Centriprep YM-30 (MILLIPORE) and dialyzed with 800 μl of 50 mM phosphate buffer (pH 5.0). The resulting solutions were used as the extracellular fraction. The yeast cells were resuspended in 50 mM phosphate buffer (pH 5.0) and were broken with a Multibeads Shocker™ (Yasui Kikai, Osaka, Japan). The crude cell extract was subjected to centrifugation at 12,000 × g for 10 min at 4 °C, and the resulting supernatant was spun at 100,000 × g for 1 h at 4 °C. The supernatant was used as the soluble fraction, and the pellet was resuspended in 300 μL of 50 mM phosphate buffer (pH 5.0) and used as the membrane fraction. PME activity was measured by a coupled spectrophotometric enzyme assay.Citation20) In this assay, formation of methanol was coupled to the reduction of NAD+, which was monitored spectrophotometrically at 340 nm. The assay mixture contained 42 mM phosphate buffer (pH 5.0), 4 mM NAD+, 1.0 U/mL of alcohol oxidase (from Pichia pastoris; WAKO), 0.35 U/mL of formaldehyde dehydrogenase (from Pseudomonas putida; TOYOBO), and 0.24% pectin (Sigma) in a total volume of 1 mL. PME-mediated methanol release is equimolar with the NADH formed in the coupled reaction. Therefore, 1 U of PME activity was defined as 1 μmol of NADH min−1 at 30 °C and pH 5.0.
PME activities of transformant strains grown on methanol are summarized in Table . The PME activities of all fractions of the wild-type strain and of the soluble fractions of strains, PMEact and PMEaod were below the limit of detection (<3 mU/mL broth) using the method described above. PME activities in the membrane and extracellular fractions of strains, PMEact and PMEaod were detected, but higher activities were observed in the extracellular fraction than in the membrane fraction, indicating that PME was successfully produced as a secreted protein. Furthermore, PME activity in the extracellular fraction of the strain PMEaod was approximately four times higher than in strain PMEact, reflecting that the AOD1 promoter was stronger than the ACT1 promoter under methanol-induced conditions.
Table 1. PME activities in the wild-type and transformant strains of C. boidinii.
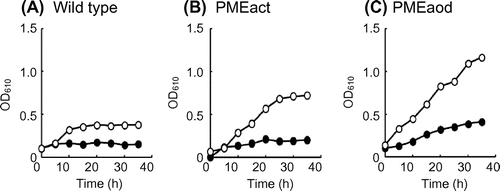
Next, we tested the growth of strains, PMEact and PMEaod on pectin (Fig. ). In the wild-type strain, the cell density obtained on pectin with a degree of methyl esterification (DE) of 90% was about two-fold higher than that obtained on pectin with a DE of 30%. This growth yield difference was due to the utilization of methyl ester moieties of pectin by the yeast. The cell densities of both strains, PMEact and PMEaod were increased compared with the wild-type strain. Furthermore, the cell density of strain PMEaod was much higher than that of strain PMEact, and the growth yield of strain PMEaod on pectin with a DE of 30% was higher than that of the wild-type strain on pectin with a DE of 90%. It has been reported that methanol-metabolizing enzymes were induced in cells grown on pectin, although the activities were lower than those in methanol-grown cells.Citation12) These results indicate that the cell density increased as the expression level of PME increased and that the PME-producing strains can utilize the methyl ester moieties in pectin more efficiently than the wild-type strain.
Fig. 1. Growth of the wild-type (A), PMEact (B), and PMEaod (C) strains on 1% pectin (DE, 30%) (●) and 1% pectin (DE, 90%) (○).
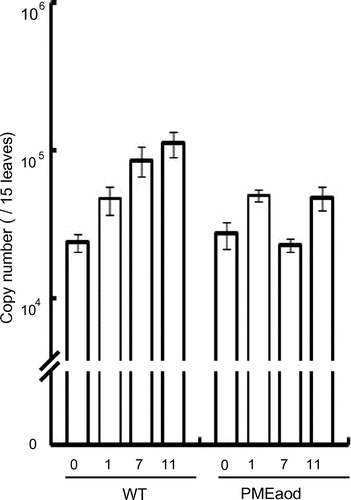
In our previous study, we demonstrated that C. boidinii cells could double 3 to 4 times on A. thaliana leaves after 11 days of inoculation.Citation10) It was also shown that methanol was the main carbon source for C. boidinii growing on A. thaliana leaves because methanol assimilation enzymes were necessary for yeast proliferation in the phyllosphere.Citation10) The PME-overproducing strain was expected to utilize the methyl ester moiety of plant pectin efficiently and to proliferate better than the wild-type strain. Therefore, we examined the effect of PME overproduction on yeast proliferation on A. thaliana leaves. We first constructed a fluorescent protein Venus-labeled variant of strain PMEaod. The expression vector pACTLV containing the Venus-encoding gene under the ACT1 promoter and LEU2 as a selectable marker was constructed using pACT1VCitation10) and pNOTeLI.Citation21) pACTLV was linearized with BglII, and introduced into C. boidinii BUL (ura3 leu2).Citation22) Furthermore, pACT1 and pAODPME were linearized with EcoRV, and used to transform this strain, yielding the Venus-labeled wild-type strain and Venus-labeled strain PMEaod. These strains were then inoculated on the leaves of A. thaliana as described previously.Citation10) To quantitate the yeast cell proliferation in the phyllosphere, quantitative PCR was performed in 20-μL mixtures in glass capillary tubes using a LightCycler (Roche Diagnostic) as described previously.Citation10) The Venus-labeled strain PMEaod was shown to survive on leaves of A. thaliana after 11 days of inoculation, although it did not proliferate on leaves as efficiently as the Venus-labeled wild-type strain (Fig. ). These results suggest that overproduction of PME negatively affected phyllospheric growth of C. boidinii. Inoculation of the wild-type strain or PME-overproducing strain on Arabidopsis did not result in any significant effects on plant growth.
Fig. 2. Quantitation of cell number of the wild-type strain and strain PMEaod on Arabidopsis leaves after 0, 1, 7, and 11 Days.
Note: Error bars show the standard deviations of triplicate measurements.
In this study, we demonstrated that overproduction of PME in C. boidinii helped cells to utilize methyl ester moieties of pectin. However, it was unable to efficiently utilize pectin on the living plant. Further study is needed to test whether the PME-producing yeast more efficiently utilizes plant biomass of dead plants because the methanol concentration on dead plant leaves was higher than that on growing plant leaves.Citation10) Although the effect of PME overproduction on yeast proliferation in the phyllosphere was negative, the present results demonstrate that heterologous proteins, such as plant growth-promoting factors, could be produced on the living plant leaves by C. boidinii. Using this method, proteins that have positive effects on plant growth can be directly produced in the phyllosphere.
Funding
This study was supported in part by a Grant-in-Aid for Scientific Research (B) [grant number 22380052 to Yasuyoshi Sakai] and a Grant-in-Aid for Scientific Research (B) [grant number 22310046], [grant number 25281063, to Hiroya Yurimoto] from the Japan Society for the Promotion of Science. It was also supported in part by Advanced Low Carbon Technology Research and Development Program (ALCA) from Japan Science and Technology Agency.
Notes
Abbreviation: PME, pectin methylesterase.
References
- MacDonald RC, Fall R. Detection of substantial emissions of methanol from plants to the atmosphere. Atmos. Environ. Part A: Gen. Top. 1993;27:1709–1713.10.1016/0960-1686(93)90233-O
- Nemecek-Marshall M, MacDonald RC, Franzen JJ, Wojciechowski CL, Fall R. Methanol emission from leaves (Enzymatic detection of gas-phase methanol and relation of methanol fluxes to stomatal conductance and leaf development). Plant Physiol. 1995;108:1359–1368.
- Fall R, Benson AA. Leaf methanol - the simplest natural product from plants. Trends Plant Sci. 1996;1:296–301.10.1016/S1360-1385(96)88175-0
- Galbally IE, Kirstine W. The production of methanol by flowering plants and the global cycle of methanol J. Atmos. Chem. 2002;43:195–229.10.1023/A:1020684815474
- Obendorf RL, Koch JL, Gorecki RJ, Amable RA, Aveni MT. Methanol accumulation in maturing seeds. J. Exp. Bot. 1990;41:489–495.10.1093/jxb/41.4.489
- Micheli F. Pectin methylesterases: cell wall enzymes with important roles in plant physiology. Trends Plant Sci. 2001;6:414–419.10.1016/S1360-1385(01)02045-3
- Ogata K, Nishikawa H, Ohsugi M. A yeast capable of utilizing methanol. Agric. Biol. Chem. 1969;33:1519–1520.10.1271/bbb1961.33.1519
- Limtong S, Srisuk N, Yongmanitchai W, Yurimoto H, Nakase T. Ogataea chonburiensis sp. nov. and Ogataea nakhonphanomensis sp. nov., thermotolerant, methylotrophic yeast species isolated in Thailand, and transfer of Pichia siamensis and Pichia thermomethanolica to the genus Ogataea. Int. J. Syst. Evol. Microbiol. 2008;58:302–307.10.1099/ijs.0.65380-0
- Peter G, Tornai-Lehoczki J, Dlauchy D. Ogataea populialbae sp. nov., a yeast species from white poplar. FEMS Yeast Res. 2009;9:936–941.10.1111/fyr.2009.9.issue-6
- Kawaguchi K, Yurimoto H, Oku M, Sakai Y. Yeast methylotrophy and autophagy in a methanol-oscillating environment on growing Arabidopsis thaliana leaves. PLoS ONE. 2011;6:e25257.10.1371/journal.pone.0025257
- Lee JD, Komagata K. Taxonomic study of methanol-assimilating yeasts. J. Gen. Appl. Microbiol. 1980;26:133–158.10.2323/jgam.26.133
- Nakagawa T, Miyaji T, Yurimoto H, Sakai Y, Kato N, Tomizuka N. A methylotrophic pathway participates in pectin utilization by Candida boidinii. Appl. Environ. Microbiol. 2000;66:4253–4257.10.1128/AEM.66.10.4253-4257.2000
- Khanh NQ, Albrecht H, Ruttkowski E, Löffler F, Gottschalk M, Jany KD. Nucleotide and derived amino acid sequence of a pectinesterase cDNA isolated from Aspergillus niger strain RH 5344. Nucleic Acids Res. 1990;18:4262.10.1093/nar/18.14.4262
- Khanh NQ, Ruttkowski E, Leidinger K, Albrecht H, Gottschalk M. Characterization and expression of a genomic pectin methyl esterase-encoding gene in Aspergillus niger. Gene. 1991;106:71–77.10.1016/0378-1119(91)90567-U
- Sakai Y, Yoshida H, Yurimoto H, Yoshida N, Fukuya H, Takabe K, Kato N. Production of fungal fructosyl amino acid oxidase useful for diabetic diagnosis in the peroxisome of Candida boidinii. FEBS Lett. 1999;459:233–237.10.1016/S0014-5793(99)01245-4
- Prodromou C, Pearl LH. Recursive PCR: a novel technique for total gene synthesis. Protein Eng. Des. Sel. 1992;5:827–829.10.1093/protein/5.8.827
- Sakai Y, Yurimoto H, Matsuo H, Kato N. Regulation of peroxisomal proteins and organelle proliferation by multiple carbon sources in the methylotrophic yeast, Candida boidinii. Yeast. 1998;14:1175–1187.10.1002/(ISSN)1097-0061
- Sakai Y, Akiyama M, Kondoh H, Shibano Y, Kato N. High-level secretion of fungal glucoamylase using the Candida boidinii gene expression system. Biochim. Biophys. Acta. 1996;1308:81–87.10.1016/0167-4781(96)00075-9
- Sakai Y, Kazarimoto T, Tani Y. Transformation system for an asporogenous methylotrophic yeast, Candida boidinii: cloning of the orotidine-5'-phosphate decarboxylase gene (URA3), isolation of uracil auxotrophic mutants, and use of the mutants for integrative transformation. J. Bacteriol. 1991;173:7458–7463.
- Grsic-Rausch S, Rausch T. A coupled spectrophotometric enzyme assay for the determination of pectin methylesterase activity and its inhibition by proteinaceous inhibitors. Anal. Biochem. 2004;333:14–18.10.1016/j.ab.2004.04.042
- Yurimoto H, Yamane M, Kikuchi Y, Matsui H, Kato N, Sakai Y. The pro-peptide of Streptomyces mobaraensis transglutaminase functions in cis and in trans to mediate efficient secretion of active enzyme from methylotrophic yeasts. Biosci. Biotechnol. Biochem. 2004;68:2058–2069.10.1271/bbb.68.2058
- Sakai Y, Tani Y. Directed mutagenesis in an asporogenous methylotrophic yeast: cloning, sequencing, and one-step gene disruption of the 3-isopropylmalate dehydrogenase gene (LEU2) of Candida boidinii to derive doubly auxotrophic marker strains. J. Bacteriol. 1992;174:5988–5993.
