Abstract
Studies in mammalian species showed that Krüppel-like factor 3 (KLF3) regulated adipose tissue development. However, it was not reported in chicken. In the current study, we found that during the growth and development of abdominal fat tissue, chicken KLF3 (Gallus gallus KLF3, gKLF3) was consecutively expressed, and its transcripts were higher at 7 weeks of age and lower at 10 weeks of age in lean broilers than in fat broilers. In addition, gKLF3 overexpression suppressed chicken CCAAT/enhancer binding protein alpha (C/EBPα), fatty acid binding protein 4 (FABP4), fatty acid synthase (FASN), and lipoprotein lipase (LPL) promoter activities, but increased chicken peroxisome proliferator-activated receptor gamma (PPARγ) promoter activity. Additionally, point mutagenesis analysis showed that the substitution of Asp by Gly within the Pro-Val-Asp-Leu-Thr (PVDLT) motif of gKLF3 significantly reduced the ability of gKLF3 to regulate the promoter activities of FABP4, FASN, LPL, C/EBPα, and PPARγ.
Graphical Abstract
Effects of Overexpression of KLF3 and Its Mutant on the Promoter Activities of Chicken FABP4, FASN, LPL, C/EBPα, and PPARγ in DF1 Cells.
Krüppel-like factors, characterized by the three C2H2 zinc fingers at the C terminus that allow sequence-specific binding to CACCC boxes and GC-rich motifs, have been shown to include at least 17 mammalian family members and to play important roles in a variety of cell processes during embryonic development and in the adult organism.Citation1)
Krüppel-like factor 3 (KLF3), a member of the Krüppel-like factor family, was first identified and cloned in murine erythroid cells.Citation2) KLF3, distinguished by its basic charge, is also known as basic Krüppel-like factor.Citation2) Mammalian KLF3 functions as a strong transcriptional repressor,Citation2,3) and its repression domain has been mapped to the N-terminal region.Citation3) The short Pro-Val-Asp-Leu-Thr (PVDLT) motif in the repression domain is critically important for its function, and it is necessary for its interaction with the well-characterized cofactor C-terminal binding protein (CtBP).Citation3) Disruption of the KLF3–CtBP interaction leads to a significant reduction in the repressive potential of KLF3.Citation3) However, loss of interaction with CtBP does not completely abolish its repression potential.Citation3) KLF3 can be modified by small ubiquitin-like modification (SUMOylation), and KLF3 SUMOylation facilitates its transcriptional repression on a composite CACCC box-GRE-driven promoter in Drosophila SL2 cells.Citation4) SUMOylation and interaction with cofactor CtBP are both critical for KLF3 function.Citation4) Lack of SUMOylation and disruption of interaction with CtBP may result in a loss of KLF3 transcription repression potential, and may also lead to a gain of transcription activation potential.Citation4)
KLF3 has been widely studied in autophagy,Citation5) erythropoiesis,Citation6) B-cell development,Citation7) myocyte differentiation,Citation8) and adipogenesis.Citation9) KLF3-knockout mice showed a marked reduction in white adipose tissue and protection from diet-induced obesity and glucose intolerance,Citation9,10) and their abdominal fat pads contained fewer and smaller cells than the wild-type littermates.Citation9) In contrast to these in vivo results, murine embryonic fibroblasts from KLF3-knockout embryos showed an increased propensity to differentiate into lipid-forming cells in vitro.Citation9) Additionally, both the mRNA and protein expression levels of KLF3 were high initially and declined as 3T3-L1 preadipocyte differentiation ensued.Citation9) Forced expression of KLF3, but not of a KLF3 mutant unable to interact with CtBP, blocked 3T3-L1 preadipocyte differentiation.Citation9) Further studies in 3T3-L1 preadipocytes showed that KLF3 bound directly to the promoter of the CCAAT/enhancer binding protein alpha (C/EBPα) gene, and suppressed C/EBPα transcription.Citation9)
Although the roles of KLF3 have been widely investigated in mammals, little is known about it in birds. The objective of the present study was to explore the gene expression pattern of KLF3 in the divergently selected chicken lines for abdominal fat content, and to explore its function in adipose tissues. Our findings provide evidence that KLF3 is involved in the regulation of chicken adipogenesis and lipid metabolism.
Materials and methods
Experimental birds and management
All animal work was conducted in accordance with the guidelines for the care and use of experimental animals established by the Ministry of Science and Technology of the People’s Republic of China (Approval number: 2006-398), and was approved by the Laboratory Animal Management Committee of Northeast Agricultural University. In total, 104 male birds, from the fourteenth generation population of Northeast Agricultural University broiler lines divergently selected for abdominal fat content (NEAUHLF; each line 52 broilers), were used. Information regarding NEAUHLF has been published previously.Citation11) Briefly, after 14 generations of divergent selection for abdominal fatness, the abdominal fat percentage of the fat broiler line at 7 weeks of age was 4.45 times as that of the lean line. All birds were kept in similar environmental conditions and had free access to food and water.
Tissues
Several male birds (each line 3–6 birds) were slaughtered at 6 h after meal at each week of age (1–12 weeks), and abdominal fat tissues were collected after slaughtering. For the birds slaughtered at 7 weeks of age, 14 other tissue samples were also collected: liver, duodenum, jejunum, ileum, pectoralis muscle, crureus muscle, gizzard, heart, spleen, kidney, pancreas, proventriculus, brain, and testis. All the tissues collected were snap-frozen and stored in liquid nitrogen until RNA extraction.
KLF3 cloning and vector construction
To clone the coding sequence (CDS) of chicken KLF3 (Gallus gallus KLF3, gKLF3), total RNA from abdominal fat tissue of 7-week-old broilers was reverse transcribed using oligo(dT)-anchor primer and ImProm-II reverse transcriptase (Promega, Madison, USA). A set of primers, gKLF3-F1 (5′-ATGAATTCTAATGGACCCCGTTTCAGTGTC-3′) and gKLF3-R1 (5′-CTCTCGAGTCAGACTAGCATGTGACGTTTTC-3′), was designed according to the predicted chicken KLF3 sequences (GenBank accession XM_427367). Additionally, ExTaq (TaKaRa, Dalian, China) was used to perform PCR reaction. The PCR conditions were as follows: pre-denaturation at 94 °C for 5 min, then 34 cycles of denaturation at 94 °C for 30 s, annealing at 62 °C for 30 s, and elongation at 72 °C for 1 min, followed by a 10 min extension at 72 °C and cooling to 4 °C. After gel extraction, the PCR product of gKLF3 was cloned into the pMD-18T vector (TaKaRa) for sequencing (BGI, Beijing, China). To generate the gKLF3 overexpression vector (pCMV-my-gKLF3), the gKLF3 cDNA fragment, produced by digestion of pMD-18T-gKLF3 using EcoRI and XhoI (TaKaRa), was subcloned into the pCMV-myc vector (Clontech, Mountain View, USA). The mut-gKLF3 overexpression plasmid, pCMV-myc-gKLF3m, with the substitution of Gly for Asp within the PVDLT sequence, was generated by the point mutation of the plasmid pCMV-my-gKLF3. The sequence CCAGTAGACCTCACG (148–162 bp, GenBank accession JX673910), encoding the PVDLT motif, was mutated into CCAGTAGGCCTCACG using the mut-primer pair (F: 5′-CCAGATGGAGCCAGTAGGCCTCACGGTGAACAAGCG-3′ and R: 5′-CGCTTGTTCACCGTGAGGCCTACTGGCTCCATCTGG-3′) and One-Tube Point Mutation Kit (Tiandz, Beijing, China).
RNA isolation and real-time PCR
Total RNA was extracted from stored tissues (100 mg) using Trizol (Invitrogen, Carlsbad, USA) following the manufacturer’s protocol. RNA quality was assessed by denaturing formaldehyde agarose gel electrophoresis. Reverse transcription (RT) was performed using 1 μg of total RNA, an oligo (dT) anchor primer (Promega), and ImProm-II reverse transcriptase (Promega). RT conditions for each cDNA amplification were 25 °C for 5 min, 42 °C for 60 min, and 70 °C for 15 min.
Real-time PCR was used to detect gene expression using the SYBR Premix Ex Taq (Takara) on a 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA) with the primers shown in Table . Chicken glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and β-actin were used as internal references. Part (1 μL) of each RT reaction product was amplified in a 20 μL PCR reaction system. Reaction mixtures were incubated in an ABI Prism 7500 sequence detection system (Applied Biosystems), programmed to conduct 1 cycle at 95 °C for 30 s and 40 cycles at 95 °C for 5 s and 60 °C for 34 s. Moreover, dissociation curves were analyzed using the Dissociation Curve 1.0 software (Applied Biosystems) for each PCR reaction to detect and eliminate possible primer–dimer artifacts. All reactions were performed in triplicate. The relative amounts of KLF3 transcripts were calculated using the comparative cycle time method.
Table 1. Primers used in RT-qPCR.
Western blot assays
Chicken DF1 cells transfected with pCMV-myc-gKLF3, pCMV-myc-gKLF3m, or pCMV-myc vector for 2 days were lysed in RIPA buffer (PBS, pH 7.4, containing 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, and a protease inhibitor cocktail). Then, the cell lysates were added into 5× denaturing loading buffer and boiled in water for 5 min. Cell lysates were separated on 5–12% SDS-polyacrylamide gels and transferred to polyvinylidenedifluoride membranes. After incubation with the primary antibody for myc-tag (1:200; Clontech), chicken GAPDH (1:1000; Beyotime, Beijing, China), chicken β-actin (1:1000; Beyotime), or chicken peroxisome proliferator-activated receptor gamma (PPARγ, described in the previous study),Citation12) a secondary horseradish peroxide-conjugated antibody (1:5000; Beyotime) was added, and a BeyoECL Plus kit (Beyotime) was used for detection.
Cell culture and luciferase assays
DF-1 chicken embryo fibroblast cell line (kindly provided by the Harbin Veterinary Research Institute, Harbin, China) and chicken preadipocytes, which were isolated according to the procedure previously described,Citation13) were grown in DMEM/F12 medium (Invitrogen), supplemented with 10% fetal calf serum (Invitrogen), and plated in 12-well dishes. In transfection assays, a fixed amount of total DNA (1 μg) was transfected in each well using the FuGENE HD transfection reagent (Roche). The transfection system for plasmids is shown in Table . After transfection and incubation for 48 h, the cells were lysed in 250 μL of 1× passive lysis buffer (Promega), and portions of the lysate were subjected to assays for firefly and renilla luciferase using the Dual-Luciferase Reporter Assay System (Promega). Promoter activity was expressed as the average of the ratio of firefly/renilla luciferase activity. All reactions were performed in triplicate.
Table 2. The transfection system of plasmids in one well.
Statistical analyses
All data are presented as means plus standard deviation (mean ± SD). Differences between groups were analyzed using unpaired two-tailed Student’s t-test. Values of p of <0.05 were considered significant unless otherwise specified.
Results
Cloning of gKLF3 gene
We cloned and sequenced the partial CDS of gKLF3, and sequence analysis showed that the acquired sequence of gKLF3 was 1005 bp long, the acquired chicken KLF3 sequence and the reference sequence (the sequence from 182 to 1186 bp, GenBank accession XM_427367.4) were identical except for two nucleotide differences, indicating that KLF3 was expressed in chicken abdominal fat tissue. These sequence data have been submitted to GenBank database under accession number JX673910.
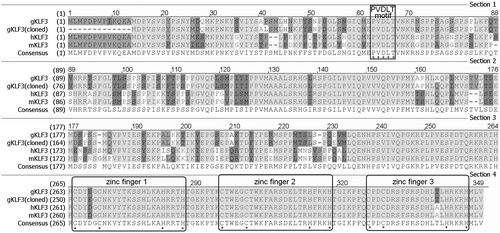
Additionally, sequence analysis showed that, the gKLF3 protein sequence (GenBank accession XP_427367.3) is closer to its human (GenBank accession NP_057615.3) and murine (GenBank accession NP_032479.1) orthologs (Fig. ).
Fig. 1. Alignment of KLF3 protein sequences.
Notes: The Align X program in Vector NTI Advance 11 (Invitrogen) was used to align the KLF3 protein sequences, gKLF3, gKLF3 (cloned), and hKLF3 and mKLF3 referred to the present chicken predicted KLF3 protein sequence (GenBank accession XP_427367.3), the KLF3 protein sequence deduced from our sequencing result (GenBank accession JX673910), the human KLF3 protein sequence (GenBank accession NP_057615.3), and the murine KLF3 protein sequence (GenBank accession NP_032479.1), respectively.
Tissue distribution of gKLF3 mRNA
Real-time RT-PCR analysis of the tissue expression pattern of gKLF3 in 7-week-old broilers from NEAUHLF showed that gKLF3 was expressed ubiquitously in chicken tissues, including duodenum, abdominal fat, brain, pectoralis muscle, proventriculus, heart, ileum, kidney, jejunum, liver, crureus muscle, gizzard, pancreas, spleen, and testis (Fig. (A)). In addition, gKLF3 was expressed at low levels in muscle tissues (pectoralis muscle and crureus muscle) and at high level in the pancreas (Fig. (A)).
Fig. 2. Tissue expression characterization of KLF3 in 7-week-old broilers.
Notes: KLF3 mRNA expression in various tissues of 7-week-old NEAUHLF male broilers (each line, n = 3) was detected by real-time RT-PCR. GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was used as a reference gene. (A) The diagrams show the relative quantification of KLF3 transcripts in a given tissue. Error bars indicate the standard deviations of six biological replicates. (B) The diagrams show the relative quantification of KLF3 transcripts in a given line and tissue. Error bars indicate the standard deviations of three biological replicates. Asterisks indicate a significant difference between fat and lean broilers, p < 0.05 (*) or p < 0.01 (**).
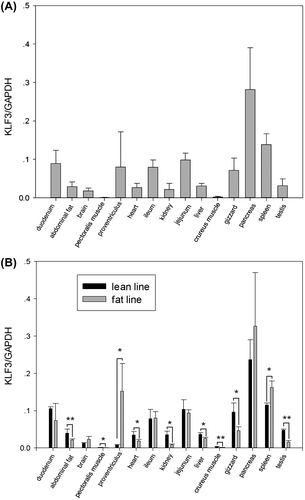
Significant differences in gKLF3 mRNA expression between fat and lean broilers were observed in 10 different tissues at 7 weeks of age. The gKLF3 transcripts were higher in proventriculus and spleen, and lower in abdominal fat, pectoralis muscle, heart, kidney, liver, crureus muscle, gizzard, and testis in fat broilers than in lean broilers (p < 0.05; Fig. (B)).
Expression pattern of gKLF3 mRNA during adipose tissue growth and development
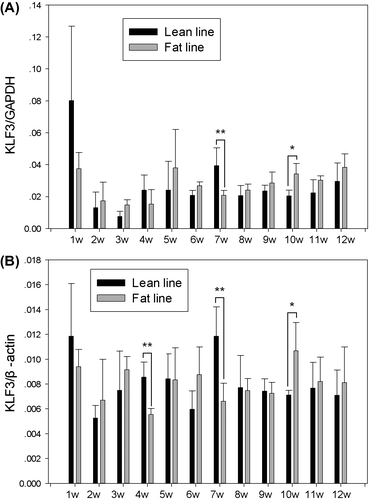
Real-time RT-PCR was used to analyze gKLF3 expression in the abdominal fat tissues of 1- to 12-week-old broilers from NEAUHLF. To avoid any possible misleading effect of using a single reference gene, two reference genes, GAPDH and β-actin, were used in real-time RT-PCR analysis. The results showed that gKLF3 was expressed in all the abdominal fat tissues tested, and its expression level (gKLF3/gGAPDH and gKLF3/gβ-actin) changed during abdominal fat tissue growth and development in both fat and lean broilers from 1 to 12 weeks of age (Fig. ). In addition, the statistical analysis showed that the relative gKLF3 mRNA expression level (gKLF3/gGAPDH) of lean males was higher at 7 weeks of age and lower at 10 weeks of age than those of fat males (p < 0.05; Fig. ). The relative gKLF3 mRNA expression level (gKLF3/gβ-actin) of lean males was higher at 4 and 7 weeks of age and lower at 10 weeks of age than those of fat broilers (p < 0.05; Fig. ). No significant difference in gKLF3 expression between fat and lean broilers was observed at the other ages tested (p > 0.05; Fig. ).
Fig. 3. Expression characterization of KLF3 in abdominal fat tissue of NEAUHLF broiler lines.
Notes: KLF3 mRNA expression in abdominal fat tissue of male broilers at various ages (each age, each line, n = 3–6) was analyzed by real-time RT-PCR. Chicken GAPDH and β-actin were used as internal controls. The diagram shows the relative quantification of KLF3 mRNA expression. Error bars indicate the standard deviation of KLF3 mRNA expression among individuals of the same age and line. Asterisks indicate significant differences between fat and lean broilers p < 0.05 (*) or p < 0.01 (**), 1–12 w = 1–12 weeks of age.
Effects of gKLF3 overexpression on promoter activities of important genes in adipose tissue
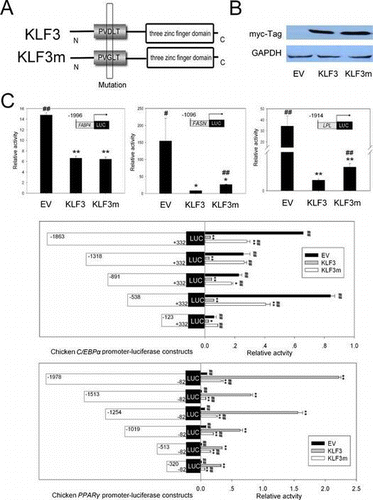
C/EBPα and PPARγ are two master positive regulators of adipogenesis in mammals and birds.Citation14–17) We investigated the effects of gKLF3 overexpression on the promoter activities of C/EBPα and PPARγ using luciferase reporter assay. As shown in Fig. , gKLF3 overexpression suppressed chicken C/EBPα promoter (−1863/+332, −1318/+332, −891/+332, −538/+332, and −123/+332) activities in DF1 cells (p < 0.05; Fig. (C)). In addition, gKLF3 overexpression also suppressed chicken C/EBPα promoter (−1863/+332) activity in the chicken preadipocytes (p < 0.01; Fig. ).
Fig. 4. Effects of KLF3 overexpression on the promoter activities of chicken FABP4, FASN, LPL, C/EBPα, and PPARγ in DF1 cells.
Notes: (A) Schematic diagram of chicken KLF3 and its mutant, KLF3m. (B) Western blot analysis of KLF3 and KLF3 mutant in DF1 cells transfected with pCMV-myc (empty vector, EV), pCMV-myc-gKLF3, and pCMV-myc-gKLF3m. (C) Effects of KLF3 and KLF3m overexpression on the promoter activities of chicken FABP4, FASN, LPL, C/EBPα, and PPARγ. Luciferase assays were conducted in DF1 cells cultured in 12-well dishes. Promoter activities are expressed as ratios of firefly/renilla luciferase activity. Asterisks indicate significant differences between the other groups and the control group, which was transfected with pCMV-myc (empty vector, EV) (*p < 0.05; **p < 0.01). The number signs indicate significant differences between the other groups and the group transfected with pCMV-myc-gKLF3 (KLF3) (#p < 0.05; ##p < 0.01). EV indicates the control group that was transfected with pCMV-myc, KLF3 indicates the group transfected with pCMV-myc-gKLF3, and KLF3m indicates the group transfected with pCMV-myc-gKLF3m.
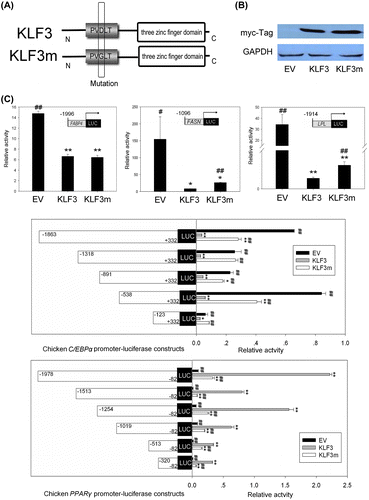
Fig. 5. Effects of KLF3 overexpression on the promoter activities of chicken C/EBPα, PPARγ, FABP4, FASN, and LPL in chicken preadipocytes.
Notes: Luciferase assays were conducted in chicken preadipocytes cultured in 12-well dishes. Promoter activities are expressed as ratios of firefly/renilla luciferase activity. Asterisks indicate significant differences between the other groups and the control group, which was transfected with pCMV-myc (empty vector, EV) (*p < 0.05; **p < 0.01). The number signs indicate significant differences between the other groups and the group transfected with pCMV-myc-gKLF3 (KLF3) (#p < 0.05; ##p < 0.01). EV indicates the control group that was transfected with pCMV-myc, KLF3 indicates the group transfected with pCMV-myc-gKLF3, and KLF3m indicates the group transfected with pCMV-myc-gKLF3m.
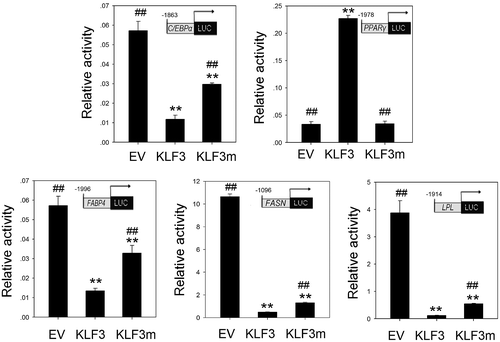
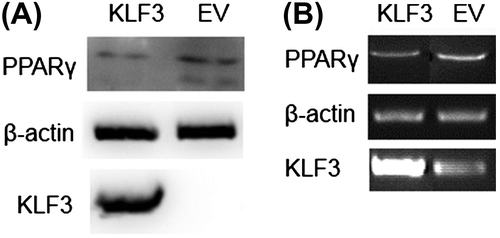
Unexpectedly, gKLF3 overexpression increased chicken PPARγ promoter (−1978/−82, −1513/−82, −1254/−82, −1019/−82, −513/−82, and −320/−82) activities in DF1 cells (p < 0.05; Fig. (C)). Additionally, gKLF3 overexpression increased chicken PPARγ promoter (−1978/−82) activities in chicken preadipocytes. However, semi-quantitative RT-PCR and western blot analysis showed that KLF3 overexpression suppressed PPARγ expression in DF1 cells (Fig. ).
Fig. 6. Effects of KLF3 overexpression on the expression of chicken PPARγ in DF1 cells.
Notes: (A) Western blot analysis of the effect of chicken KLF3 overexpression on the expression of PPARγ in DF1 cells. (B) Semi-quantitative RT-PCR analysis of the effect of chicken KLF3 overexpression on the expression of PPARγ in DF1 cells. EV indicates the control group that was transfected with pCMV-myc, and KLF3 indicates the group transfected with pCMV-myc-gKLF3.
Adipose tissue plays an important role in lipid metabolism. To assess whether gKLF3 regulates lipid metabolism in adipose tissue, we also investigated the effect of gKLF3 overexpression on promoter activities of chicken fatty acid binding protein 4 (FABP4), fatty acid synthase (FASN), and lipoprotein lipase (LPL) genes. As shown in Figs. and , gKLF3 overexpression significantly suppressed the promoter activities of chicken FABP4 (−1996/+22), FASN (−1096/+160), and LPL (−1914/+66) in DF1 cells and chicken preadipocytes (Figs. (C) and ; p < 0.05).
Effect of a single mutation of the Asp residue in the PVDLT motif on the transcriptional regulatory capacity of KLF3
A previous study in mammalian species showed that substitution of the two consecutive residues Asp-Leu for Ala-Ser or Ala-Ala in the PVDLT motif significantly compromised the ability of KLF3 to repress transcription.Citation3,4) However, the effect of the mutation of the residue Asp in the PVDLT motif on KLF3 activitie remains unclear. In the present study, we generated a chicken KLF3 mutant (KLF3m) with a mutation of Asp to Gly in the PVDLT motif using point mutagenesis (Fig. (A)). The luciferase assay showed that similar to wild type chicken KLF3 (gKLF3), gKLF3m overexpression (Fig. (B)) also significantly suppressed the promoter activities of chicken FABP4 (−1996/+22), FASN (−1096/+160), LPL (−1914/+66), and C/EBPα (−1863/+332) in both DF1 cells (p < 0.05, Fig. (C)) and chicken preadipocytes (p < 0.05, Fig. ), and increased chicken PPARγ promoter (−1978/−82, −1513/−82, −1254/−82, −1019/−82, −513/−82, and −320/−82) activities in DF1 cells (p < 0.05, Fig. (C)). However, compared with gKLF3 overexpression, gKLF3m overexpression showed reduced repression ability of promoter activities of chicken FABP4 (−1996/+22), FASN (−1096/+160), LPL (−1914/+66), and C/EBPα (−1863/+332, −1318/+332, −891/+332, −538/+332, and −123/+332), and reduced activation of promoter activities of chicken PPARγ (−1978/−82, −1513/−82, −1254/−82, −1019/−82, −513/−82, and −320/−82) in DF1 cells (p < 0.05, Fig. (C)) or chicken preadipocytes (p < 0.05, Fig. ).
Interestingly, significant difference in repression of chicken FABP4 promoter (−1996/+22) between gKLF3m and gKLF3 overexpression was observed in chicken preadipocytes (p < 0.05, Fig. ), but not in DF1 cells (p > 0.05, Fig. (C)). In addition, unlike in DF1 cells, no significant effect of gKLF3m overexpression on chicken PPARγ (−1978/−82) promoter activity was observed in chicken preadipocytes (p > 0.05, Fig. ).
Discussion
KLFs are a group of ancient and conserved members of the multiple zinc-finger transcription factors. Previous studies have showed that several members of the KLF family play roles in the mammalian adipogenesis and lipogenesis.Citation14,16,18) Our previous study indicated that one member of KLFs, KLF7, is expressed in chicken adipose tissues, and plays a negative role in the regulation of chicken adipogenesis.Citation13,19) In the current study, the results showed that KLF3, an another member of KLFs, was also expressed in chicken adipose tissue. In addition, the sequence alignment showed that KLF3 protein is highly conserved among chicken, mouse and human, suggesting that KLF3 may have a similar function in these species (Fig. ).
The tissue expression analysis showed that chicken, human, and mouse KLF3s have similar expression patterns.Citation2,20) The only difference was that, unlike human KLF3,Citation20) gKLF3 was expressed at high level in the pancreas (Fig. (A)).
Comparision of gKLF3 transcripts in 15 tissues from fat and lean broilers at 7 weeks of age showed that the gKLF3 transcripts were higher in proventriculus and spleen, and lower in abdominal fat, pectoralis muscle, heart, kidney, liver, crureus muscle, gizzard, and testis in fat broilers than in lean broilers (p < 0.05; Fig. (B)).
In the current study, real-time RT-PCR analysis showed that gKLF3 was expressed consecutively during the chicken adipose tissue development from 1 to 12 weeks of age, and its expression levels clearly changed during abdominal fat tissue development in both the fat and lean broilers, indicating that, consistent with the report in mouse,Citation9) gKLF3 might also regulate the growth and development of chicken adipose tissue. In addition, the relative gKLF3 mRNA expression levels of lean males were higher at 7 weeks of age and lower at 10 weeks of age than those of fat males, suggested that the difference of KLF3 expression in abdominal adipose tissue might contribute to the fatness trait difference between fat and lean birds.
C/EBPα and PPARγ are two master positive regulators of adipogenesis in mammals and birds.Citation14–17) In the current study, we found that gKLF3 overexpression suppressed chicken C/EBPα promoter activity in DF1 cells and chicken preadipocytes (p < 0.01; Fig. ). This is consistent with the report in 3T3-L1 preadipocytes that KLF3 overexpression suppressed C/EBPα expression.Citation9) In addition, KLF3-induced suppression of C/EBPα promoter was observed in all five different lengths of C/EBPα promoter in DF1 cells, suggesting that, similar to murine KLF3,Citation9) the gKLF3 binding site might be also located close to the transcription start site of chicken C/EBPα.
Unexpectedly, we found that gKLF3 overexpression increased chicken PPARγ promoter activity in DF1 cells and chicken preadipocytes (p < 0.01; Fig. ). These results differ from the previous studies in the 3T3-L1 cell system that KLF3 overexpression suppressed PPARγ expression.Citation9) However, we indeed found that KLF3 overexpression suppressed PPARγ expression in DF1 cells using RT-qPCR and western blot (Fig. ). The discrepancy between results of reporter gene assay and semi-quantitative RT-PCR and western blot analyses may be due to the limitations of reporter gene assay: the structure of PPARγ promoter on the luciferase vector was not the same as that on the chromosome. In addition, the PPARγ promoter on the luciferase vector does not contain histones, and is not effected by epigenetics modification. Together, these results indicated that KLF3 may play an essential role in chicken adipogenesis, at least in part through regulating the expression of C/EBPα and PPARγ.
FASN is involved in de novo fatty acid synthesis, which occurs primarily in the liver and adipose tissue, by catalyzing the synthesis of palmitate from acetyl-CoA and malonyl-CoA into long-chain saturated fatty acids.Citation21) Herein, the suppression of FASN promoter activity by KLF3 indicated that, besides the role of maintaining normal fatty acid composition by regulating genes involved in a fatty acid desaturation pathway revealed in C. elegans,Citation22) KLF3 might also inhibit de novo fatty acid synthesis in chicken.
Triglyceride (TG) is hydrophobic and cannot be transported in circulation; it is therefore assembled into a blend of lipid and protein termed lipoproteins, which can transport hydrophobic molecules. FABP4 is a cytosolic protein that functions as a lipid chaperone in adipocytes.Citation23) LPL is a protein that plays a central role in plasma TG metabolism by hydrolyzing TG-rich chylomicrons and very low-density lipoproteins. KLF3 overexpression-mediated suppression on the promoter activities of FABP4 and LPL indicated that, consistent with the report in C. elegans,Citation24) gKLF3 suppressed lipoprotein assembly and mobilization.
Taken together, there would be two possible ways for gKLF3 to regulate lipid metabolism in chicken. First, gKLF3 directly suppresses lipid biosynthesis by inhibiting FASN transcription. Alternatively, gKLF3 represses lipid circulation by suppressing the expression of FABP4 and LPL.
In addition, our results also showed that compared with gKLF3 overexpression, the overexpression of gKLF3m, a mutated gKLF3 with a mutation of Asp to Gly in the PVDLT motif (Fig. (B)), had reduced suppression of the promoter activities of chicken FABP4, FASN, LPL, and C/EBPα both in DF1 cells and chicken preadipocytes, and reduced activation of chicken PPARγ promoter activities in DF1 cells (p < 0.05, Fig. (C)). These results indicated that the mutation of Asp to Gly in the PVDLT motif reduced, but did not abolish, KLF3 activity. This is consistent with the results of double mutation of Asp-Leu to Ala-Ser or Ala-Ala in the PVDLT motif,Citation3,4) indicating that the Asp is the key residue in the PVDLT sequence.
Collectively, the current study demonstrated that chicken KLF3 is expressed in chicken adipose tissue, and regulates important genes involved in adipogenesis and lipid metabolism; and the mutation of the Asp in PVDLT significantly compromise, but does not abolish, KLF3 activity.
Funding
This study was supported by the National Basic Research Program of China [grant number 2009CB941604]; National 863 Project of China [grant number 2013AA102501]; China Agriculture Research System [grant number CARS-42]; the Program for the Innovation Research Team at the University of Heilongjiang Province [grant number 2010td02]; and the Program for High-level personnel start at Shihezi University [grant number RCZX201334].
References
- Suske G, Bruford E, Philipsen S. Mammalian SP/KLF transcription factors: bring in the family. Genomics. 2005;85:551–556.10.1016/j.ygeno.2005.01.005
- Crossley M, Whitelaw E, Perkins A, Williams G, Fujiwara Y, Orkin SH. Isolation and characterization of the cDNA encoding BKLF/TEF-2, a major CACCC-box-binding protein in erythroid cells and selected other cells. Mol. Cell. Biol. 1996;16:1695–1705.
- Turner J, Crossley M. Cloning and characterization of mCtBP2, a co-repressor that associates with basic Kruppel-like factor and other mammalian transcriptional regulators. EMBO J. 1998;17:5129–5140.10.1093/emboj/17.17.5129
- Perdomo J, Verger A, Turner J, Crossley M. Role for SUMO modification in facilitating transcriptional repression by BKLF. Mol. Cell. Biol. 2005;25:1549–1559.10.1128/MCB.25.4.1549-1559.2005
- Guo L, Huang JX, Liu Y, Li X, Zhou SR, Qian SW, Liu Y, Zhu H, Huang HY, Dang YJ, Tang QQ. Transactivation of Atg4b by C/EBP promotes autophagy to facilitate adipogenesis. Mol. Cell. Biol. 2013;33:3180–3190.10.1128/MCB.00193-13
- Funnell AP, Maloney CA, Thompson LJ, Keys J, Tallack M, Perkins AC, Crossley M. Erythroid Kruppel-like factor directly activates the basic Kruppel-like factor gene in erythroid cells. Mol. Cell. Biol. 2007;27:2777–2790.10.1128/MCB.01658-06
- Ulgiati D, Subrata LS, Abraham LJ. The role of Sp family members, basic Kruppel-like factor, and E box factors in the basal and IFN-gamma regulated expression of the human complement C4 promoter. J. Immunol. 2000;164:300–307.
- Himeda CL, Ranish JA, Pearson RC, Crossley M, Hauschka SD. KLF3 regulates muscle-specific gene expression and synergizes with serum response factor on KLF binding sites. Mol. Cell. Biol. 2010;30:3430–3443.10.1128/MCB.00302-10
- Sue N, Jack BH, Eaton SA, Pearson RC, Funnell AP, Turner J, Czolij R, Denyer G, Bao S, Molero-Navajas JC, Perkins A, Fujiwara Y, Orkin SH, Bell-Anderson K, Crossley M. Targeted disruption of the basic Kruppel-like factor gene (Klf3) reveals a role in adipogenesis. Mol. Cell. Biol. 2008;28:3967–3978.10.1128/MCB.01942-07
- Bell-Anderson KS, Funnell AP, Williams H, Mat Jusoh H, Scully T, Lim WF, Burdach JG, Mak KS, Knights AJ, Hoy AJ, Nicholas HR, Sainsbury A, Turner N, Pearson RC, Crossley M. Loss of Kruppel-like factor 3 (KLF3/BKLF) leads to upregulation of the insulin-sensitizing factor adipolin (FAM132A/CTRP12/C1qdc2). Diabetes. 2013;62:2728–2737.10.2337/db12-1745
- Guo L, Sun B, Shang Z, Leng L, Wang Y, Wang N, Li H. Comparison of adipose tissue cellularity in chicken lines divergently selected for fatness. Poult. Sci. 2011;90:2024–2034.10.3382/ps.2010-00863
- Wang L, Na W, Wang Y, Wang Y, Wang N, Wang Q, Li Y, Li H. Characterization of chicken PPARγ expression and its impact on adipocyte proliferation and differentiation. Yichuan. 2012;34:76–86.
- Zhang Z, Wang H, Sun Y, Li H, Wang N. Klf7 modulates the differentiation and proliferation of chicken preadipocyte. Acta Biochim. Biophys. Sin. 2013;45:280–288.10.1093/abbs/gmt010
- Rosen ED, MacDougald OA. Adipocyte differentiation from the inside out. Nat. Rev. Mol. Cell. Biol. 2006;7:885–896.10.1038/nrm2066
- Wang Y, Mu Y, Li H, Ding N, Wang Q, Wang Y, Wang S, Wang N. Peroxisome proliferator-activated receptor-gene: a key regulator of adipocyte differentiation in chickens. Poult. Sci. 2008;87:226–232.10.3382/ps.2007-00329
- Farmer SR. Transcriptional control of adipocyte formation. Cell Metab. 2006;4:263–273.10.1016/j.cmet.2006.07.001
- Liu S, Wang Y, Wang L, Wang N, Li Y, Li H. Transdifferentiation of fibroblasts into adipocyte-like cells by chicken adipogenic transcription factors. Comp. Biochem. Physiol. A: Mol. Integr. Physiol. 2010;156:502–508.10.1016/j.cbpa.2010.04.003
- Lefterova MI, Lazar MA. New developments in adipogenesis. Trends Endocrinol. Metab. 2009;20:107–114.10.1016/j.tem.2008.11.005
- Zhang ZW, Wang ZP, Zhang K, Wang N, Li H. Cloning, tissue expression and polymorphisms of chicken Krüppel-like factor 7 gene. Anim. Sci. J. 2013;84:535–542.10.1111/asj.2013.84.issue-7
- Wang MJ, Qu XH, Wang LS, Zhai Y, Wu SL, He FC. cDNA cloning, subcellular localization and tissue expression of a new human Kruppel-like transcription factor: human basic Kruppel-like factor (hBKLF). Yi Chuan Xue Bao. 2003;30:1–9.
- Smith S, Witkowski A, Joshi AK. Structural and functional organization of the animal fatty acid synthase. Prog. Lipid Res. 2003;42:289–317.10.1016/S0163-7827(02)00067-X
- Zhang J, Yang C, Brey C, Rodriguez M, Oksov Y, Gaugler R, Dickstein E, Huang CH, Hashmi S. Mutation in Caenorhabditis elegans Krüppel-like factor, KLF-3 results in fat accumulation and alters fatty acid composition. Exp. Cell Res. 2009;315:2568–2580.10.1016/j.yexcr.2009.04.025
- Makowski L, Hotamisligil GS. The role of fatty acid binding proteins in metabolic syndrome and atherosclerosis. Curr. Opin. Lipidol. 2005;16:543–548.10.1097/01.mol.0000180166.08196.07
- Zhang J, Hashmi S, Cheema F, Al-Nasser N, Bakheet R, Parhar RS, Al-Mohanna F, Gaugler R, Hussain MM, Hashmi S. Regulation of lipoprotein assembly, secretion and fatty acid β-oxidation by Krüppel-like transcription factor, klf-3. J. Mol. Biol. 2013;425:2641–2655.10.1016/j.jmb.2013.04.020
