Abstract
We developed an efficient screening method for Saccharomyces cerevisiae strains from environmental isolates. MultiPlex PCR was performed targeting four brewing S. cerevisiae genes (SSU1, AWA1, BIO6, and FLO1). At least three genes among the four were amplified from all S. cerevisiae strains. The use of this method allowed us to successfully obtain S. cerevisiae strains.
Saccharomyces cerevisiae is one of the most important microbes in fermentation industries. Various yeasts exist in the natural environment, with some showing similar properties to S. cerevisiae in terms of ethanol production, flavor, cell size, and colony. It is therefore necessary to isolate true S. cerevisiae strains suitable for brewing.Citation1) Here, we developed an efficient screening method for S. cerevisiae using gene-specific Multiplex PCR.
We first selected four highly conserved genes (SSU1, AWA1, BIO6, and FLO1) in brewing S. cerevisiae strains. SSU1 encodes a plasma membrane protein, which is involved in sulfite resistance.Citation2) AWA1 encodes a serine- and threonine-rich glycosylphosphatidylinositol-anchored protein that is localized to the cell wall. Citation3,4) BIO6 is involved in biotin synthesis,Citation5) though sake yeasts do not require biotin for growth. FLO1 is responsible for flocculation of S. cerevisiae.Citation6,7) These genes are not only highly conserved in industrial S. cerevisiae, but their products are also preferable for brewing.
To determine the presence of the four genes in S. cerevisiae, we performed Multiplex PCR using genomic DNA of various S. cerevisiae strains, listed in Table . The primer pairs used in this study are listed in supplemental table (see Biosci. Biotechnol. Biochem. Web site). The unknown yeast strains were isolated from environmental samples. Yeast strains were cultivated in YPD medium (1% yeast extract, 2% peptone, and 2% dextrose) without shaking at 30 °C. Genomic DNAs were obtained using the MagExtractor® Genome (TOYOBO) according to the manufacturer’s instructions. DNA concentrations were measured using the NanoDrop2000 (Thermo Fisher Science) and adjusted to 1 μg/mL. Multiplex PCR was performed according to the manufacturer’s instructions (Takara). One nanogram DNA was used per 5 μL Multiplex PCR reaction mixture. Amplification of DNA was carried out for 40 cycles under the following conditions: denaturation, 94 °C for 1 min; primer annealing, 53 °C for 90 s; primer extension, 72 °C for 2 min. Each primer pair was added at a final concentration of 0.2 μM. The electrophoresis on a 2% agarose gel detected correct sizes of the two to four amplified DNA bands from almost all S. cerevisiae strains with some exceptions as follows: SSU1 gene in lane 14; AWA1 gene in lanes 6, 7, 8, 10, 11, 14, 15, 16, and 17; BIO6 gene in lanes 6, 8, 10, 15, 16, and 17. The amplified DNA bands were confirmed of the corresponding genes by sequencing analysis. In contrast, when Multiplex PCR analysis was performed using genomic DNAs of the non-S. cerevisiae strains (Table ), the amplified DNA bands’ corresponding genes were not detected (Fig. (B)). Only in lane 22, the DNA band was detected clearly; however, the size of DNA band was not correct and subsequent sequencing analysis revealed that this DNA was not corresponding to these four genes. These results indicate that more than two genes among the above four genes are present, and that the strains may be candidate S. cerevisiae strains. And Multiplex PCR targeting the four genes is effective and sufficient for the screening of S. cerevisiae strains.
Table 1. Yeast strains used in this study.
Fig. 1. Multiplex PCR analysis of SSU1, AWA1, BIO6, and FLO1 using DNA of various S. cerevisiae strains (A), other genera (B), and environmental isolates (C).
Note: Arrows I, II, III, and IV indicate DNA bands corresponding to SSU1 (1049 bp), AWA1 (898 bp), BIO6 (595 bp), and FLO1 (315 bp), respectively. Lane numbers correspond to the strain list numbers in Table .
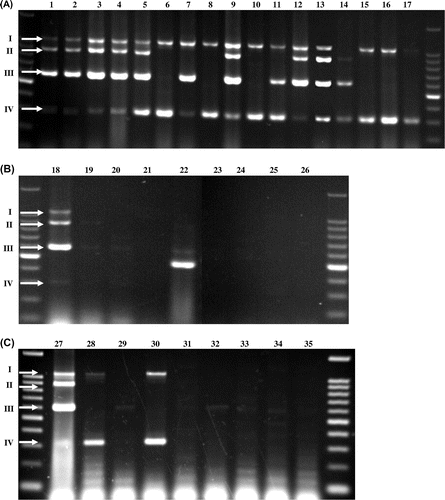
We subsequently applied this method to environmental yeast isolates. As shown in Fig. (C), certain electrophoresis profiles were observed; two bands corresponding to SSU1 and FLO1 were detected in lanes 28 and 30, respectively, a single band corresponding to BIO6 in lane 32, and one band in lane 29 not corresponding to any of the four genes. Numerous DNA bands were detected in lanes 31, 33, 34, and 35. The 28S rRNA gene sequences of these isolates were highly similar to S. cerevisiae (28 and 30), Candida sp. (29 and 32), and Pichia sp. (31, 33, 34, and 35). These two S. cerevisiae strains did not show high ethanol production abilities compared with K7 or K9.
Yamagishi et al.Citation8) and Shimizu et al.Citation4) previously reported that brewing and non-brewing yeasts are distinguishable using restriction fragment length polymorphism of the FLO1 gene and amplified fragment length polymorphism of the AWA1 gene. In this study, we found that Multiplex PCR targeting four genes (SSU1, AWA1, BIO6, and FLO1) is efficient to screen brewing S. cerevisiae strains. We applied this method to 150 yeast isolates from various environmental samples. All four genes were amplified from 20 isolates, with 5 isolates lacking FLOI. Subsequent sequencing analyses of 28S rRNA gene revealed that all 25 strains were identified as S. cerevisiae (Table ), and all exhibited high ethanol production abilities (3–6% ethanol concentration) compared with K7 and K9 strains with YPD medium (1% Yeast extract, 2% peptone, and 10% dextrose) by gas chromatography. Sequencing analyses of the 28S rRNA gene of 20 strains selected at random in other 125 strains revealed that 3 strains identified as Pichia, 7 strains identified as Candida, and 10 strains identified as Torulaspora. Using this method before identification by sequencing analysis is expected to reduce the time and cost when isolating the novel S. cerevisiae strains from the environment. In this study, we could not observe the phenotypes corresponding to the genes; however, we could isolate S. cerevisiae strains, which exhibit high ethanol production abilities compared with Kyokai strains, by the Multiplex PCR method using these four genes.
Table 2. Occurrence of Four Genes and Identification of Environmental Isolates.
Supplemental material
The supplemental material for this paper is available online at http://dx.doi.10.1080/09168451.2014.910098.
Supplemental Table 1
Download PDF (93.7 KB)References
- Kodama K. Sake-brewing yeasts. In: Rose AH, Harrison JS, editors. The yeasts. 2nd ed. Vol. 5. London: Academic Press; 1993. p. 129–168.
- Park H, Bakalinsky AT. SSU1 mediates sulphite efflux in Saccharomyces cerevisiae. Yeast. 2000;16:881–888.10.1002/(ISSN)1097-0061
- Shimoi H, Sakamoto K, Okuda M, Atthi R, Iwashita K, Ito K. The AWA1 gene is required for the foam forming phenotype and cell surface hydrophobicity of sake yeast. Appl. Environ. Microbiol. 2002;68:1932–1937.
- Shimizu M, Miyashita K, Kitagaki H, Ito K, Shimoi H. Amplified fragment length polymorphism of the AWA1 gene of sake yeasts for identification of sake yeasts strains. J. Biosci. Bioeng. 2005;100:678–680.10.1263/jbb.100.678
- Wu H, Ito K, Shimoi H. Identification and characterization of a novel biotin biosynthesis gene in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2005;71:6845–6855.10.1128/AEM.71.11.6845-6855.2005
- Miki BL, Poon NH, James AP, Seligy VL. Possible mechanism for flocculation interactions governed by gene FlO1 in Saccharomyces cerevisiae. J. Bacteriol. 1982;150:879–889.
- Govender P, Domingo JL, Bester MC, Pretorius IS, Bauer F. Controlled expression of the dominant flocculation genes FLO1, FLO5, and FLO11 in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2008;74:6041–6052.10.1128/AEM.00394-08
- Yamagishi H, Otuta Y, Funahashi W, Ogata T, Sakai K. Differentiation between brewing and non-brewing yeasts using a combination of PCR and RFLP. J. Appl. Microbiol. 1999;86:505–513.10.1046/j.1365-2672.1999.00691.x
