Abstract
Phospholipase D (PLD) catalyzes the hydrolysis of phosphatidylcholine (PC), the most abundant phospholipids of plasma membrane, resulting in the production of choline and phosphatidic acid (PA). Choline is a precursor of the neurotransmitter acetylcholine, whereas PA functions as an intracellular lipid mediator of diverse biological functions. For assessing PLD activity in vitro, PLD-derived choline has been often analyzed with radioactive or non-radioactive methods. In this study, we have developed a new method for detecting choline and PA with MALDI-QIT-TOF/MS by using 9-aminoacridine as a matrix. The standard calibration curves showed that choline and PA could be detected with linearity over the range from 0.05 and 1 pmol, respectively. Importantly, this method enables the concomitant detection of choline and PA as a reaction product of PC hydrolysis by PLD2 proteins. Thus, our simple and direct method would be useful to characterize the enzymatic properties of PLD, thereby providing insight into mechanisms of PLD activation.
Graphical Abstract
9-aminoacridine is a matrix with low background and high sensitivity to analyze PA and choline by MALDI-QIT-TOF/MS.
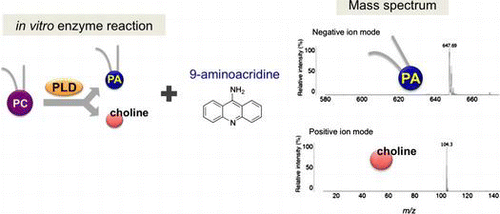
Phosphatidylcholine (PC) is the most abundant phospholipids representing 50% of cellular membrane in eukaryotic cells.Citation1) The cellular amount of PC, which is determined by a balance of its synthesis and hydrolysis, plays an important role for cell proliferation, survival, apoptosis, and maintenance of homeostasis.Citation2–4) Phospholipase D (PLD), which is broadly distributed in bacteria, fungi, plants, and animals, is known to hydrolyze the phosphodiester bond between the phosphate and choline of PC, resulting in the production of free choline and phosphatidic acid (PA).Citation5) Importantly, the PLD-derived choline is an essential precursor of the neurotransmitter acetylcholine, while PA acts as a lipid second messenger that controls the actin cytoskeleton, vesicle trafficking for secretion and endocytosis, and receptor signaling.Citation6,7) Among mammalian PLD family members, PLD2 protein is often used for in vitro PLD activity assay, because PLD2 exhibits constitutive activity.Citation8)
There are several methods for assessing PLD activity in vitro. The first is the classical thin-layer chromatography (TLC), in which [3H]PC is incubated with purified PLD protein, and the reaction products containing radio labeled-choline are separated in an aqueous phase.Citation9) Another method is a non-radioactive technique, which requires two steps for enzymatic processes in addition to PLD reaction; choline quantification by using choline oxidase that produces betaine and hydrogen peroxide from choline components as substrate. In the presence of horseradish peroxidase, hydrogen peroxide reacts with Amplex Red reagent, which in turn generate the highly fluorescent resorufin.Citation10) In contrast to the above, a three-step enzymatic reaction method is to detect PC-derived PA by using PA-specific lipase that hydrolyzes PA to two fatty acids and glycerol-3-phosphate (G3P). Subsequently, hydrogen peroxide is generated by G3P oxidase followed by quantification using horseradish peroxidase with the Amplex Red system.Citation11) Although these methods have been widely employed to measure PLD activity due to their simplicity and sensitivity, there is no way to assess PLD activity by directly detecting both choline and PA.
In recent years, matrix-assisted laser desorption/ionization-quadrupole ion trap-time-of-flight mass spectrometry (MALDI-QIT-TOF/MS) has been used as a novel technique for lipid analysis.Citation12,13) The choice of matrix used for MALDI-TOF/MS depends on the mass range and chemical properties of analytes. Among many kinds of matrices, sinapinic acid is generally used for high-weight molecules such as proteins,Citation14,15) while α-cyano-4-hydroxycinnamic acid (CHCA) is often used for middle weight molecules such as peptides.Citation16) 2,5-dihydroxybenzoic acid (DHBA) or 9-aminoacridine (9-AA) is generally used for low molecular weight molecules, such as lipids or metabolites.Citation15,17,18) However, strong background interferences are observed when CHCA as matrix was used in the low mass ranges under positive ion mode. Recently, it was reported that 9-AA provides higher sensitivity than DHBA and that reduced the matrix background as a competitor for protons in MS spectrum.Citation19–21)
In this study, we have established a simple, sensitive, and rapid method for detecting choline and PA with MALDI-QIT-TOF/MS. By using 9-AA as a matrix, standard molecules of choline and PA are predominantly detected with positive and negative ion modes, respectively. Furthermore, we demonstrated that 9-AA can be used to detect PLD-derived choline and PA followed by hydrolysis of PC. Our data present the single-step method that allows for the evaluation of PLD activity by directly and simultaneously detecting choline and PA with MALDI-QIT-TOF/MS.
Materials and methods
Materials
All chemicals were of the highest purity obtainable. DHBA and CHCA were purchased from Shimadzu (Kyoto, Japan). 9-aminoacridine hemihydrate (9-AA) was purchased from Acros Organics (Morris Plains, NJ, USA). Choline hydroxide solution (46 wt. % in H2O) was purchased from Sigma-Aldrich Japan (Tokyo, Japan). 1,2-dipalmitoyl-sn-glycero-3-phosphatidic acid, disodium salt (DPPA), and 1,2-dipalmitoyl-sn-glycero-3-phosphatidylcholine disodium salt (DPPC) were purchased from Wako Pure Chemical (Osaka, Japan). [methyl-3H]choline chloride, [choline-methyl-3H]dipalmitoyl-PC, and DPPC (1 mCi/mL) were purchased from Perkin–Elmer Life and Analytical Sciences (Boston, MA, USA). Streptomyces sp. PLD and 5-fluoro-2-indoly des-chlorohalopemide (FIPI) were purchased from Cayman Chemical (Ann Arbor, MI, USA). Merck HPTLC plates (Silica gel 60 Å, 20 × 10 cm, glass plates) were purchased from Millipore Japan (Tokyo, Japan). Bradykinin fragment 1–7, the calibration standard for MALDI-QIT-TOF/MS, was purchased from Sigma-Aldrich Japan (Tokyo, Japan).
MALDI-QIT-TOF/MS and dissociation profiles
DHBA, CHCA or 9-AA matrix (0.4 mg) was dissolved in 50 mL of 90% methanol. Samples dissolved in 90% methanol (0.5 mL) and matrix solution (0.5 mL) were deposited on a MALDI plate and left to dry at room temperature to prepare sample spots. External calibrations were achieved using the standard reagents of DHBA matrix (monoisotopic mass of [(M + H)+ = 155.03] and bradykinin fragment 1–7 (monoisotopic mass of [(M + H)+ = 757.4]). Mass spectra were acquired using a MALDI-QIT-TOF/MS (AXIMA-Resonance, Shimadzu, Kyoto, Japan). Samples were irradiated by a nitrogen UV laser (337 nm). Helium gas was used for ion cooling in the ion trap, and argon gas was used for collision induced dissociation (CID) fragmentation. Each mass spectrum represented the accumulation of 400 laser shots (sum of 2 × 200). This was repeated for the other two sample spots. Peak intensity of spectral mass resolutions and signal-to-noise ratio of 3:1 were determined by the software for the instrument “AXIMA launchpad 2.9” (Shimadzu, Kyoto, Japan). CID spectra were acquired using an activation time of 30 ms, and varying collision energies of up to 300 (arbitrary unit given by the instrument; ranges from zero to 1000) using argon as collision gas. The percent intensity of the precursor and major product ions relative to their total ion intensity were plotted against the collision energy. The ion source pressure during MS/MS measurements was typically 2.5 × 10−6 Pa for the reflectron part and 5 × 10−5 Pa inside the QIT using helium for collisional ion cooling. Each profile in MS and MS/MS mode was the result of 400 single laser shots (sum of 2 × 200) directed onto the selected sample preparation, which were accumulated to give the final mass spectrum or CID spectrum.
Preparation of standard choline and PA for determining detection limits
Choline hydroxide was diluted from 2 × 10−9 to 2 × 10−3 M and DPPA was diluted from 2 × 10−8 mM to 2 × 10−3 M using the stock solution in 90% methanol, respectively. 0.5 μL of analytes was mixed with 0.5 μL of 9-AA matrix solution (20 mg/mL in 90% methanol) on a MALDI plate.
Plasmids
Mouse PLD2 wild-type and K758R mutant cDNAs (generously gifted from Dr. Yasunori Kanaho, University of Tsukuba, JAPAN) were tagged at the C terminus with hemagglutinin (HA) sequences and cloned into the expression vector pcDNA3 (Invitrogen, Carlsbad, CA, USA) to produce pcDNA3-PLD2(WT)-HA and pcDNA3-PLD2(K758R)-HA.
Cell culture and transfection
The human embryonic kidney (HEK) 293T cells were maintained in Dulbecco’s modified Eagle’s medium (Wako Pure Chemical Industries, Ltd. Osaka, Japan) supplemented with 10% fetal bovine serum, penicillin (100 units/mL), streptomycin (100 μg/mL) at 37 °C in 5% CO2. HEK293T cells were transfected with GeneJuice Transfection Reagent (Novagen, Darmstadt, Germany) according to the manufacturer’s protocol.
Immunoprecipitation and Western blot
Transfected HEK293T cells with pcDNA3-HA, PLD2 (WT)-HA, and PLD2 (K758R)-HA expression plasmids were incubated for 36 h. After incubation, cells were washed in ice-cold PBS, and resuspended in lysis buffer (20 mM Hepes-NaOH (pH 8.0), 150 mM NaCl, 1% NP-40, 1 mM EDTA, and 1× protease inhibitors (nacalai tesque, Kyoto, Japan)). After 30 min on ice, lysates were cleared by centrifugation at 14,000 rpm for 10 min at 4 °C. The supernatant was treated with anti-HA antibody (12CA5, Roche Applied Science). Immunoprecipitated proteins resolved by 10% SDS-PAGE were transferred to an Immobilon-P membrane (Millipore Japan, Tokyo, Japan) before Western blot detection using anti-HA antibody (3F10, Roche Applied Science) at 1:2000 dilution. Visualization was carried with horseradish peroxidase-coupled secondary antibody and developed using Luminata Forte Western HRP substrate (Millipore Japan, Tokyo, Japan) and exposed to medical X-ray films (Fuji Film, Kanagawa, Japan).
In vitro PLD assay for TLC
PLD activity was determined by detection of the generated 3H-labeled choline from [choline-methyl-3H] dipalmitoyl-PC as previously described.Citation22,23) For the in vitro PLD assay, the immunoprecipitated proteins were incubated with 3 μL of [choline-methyl-3H] dipalmitoyl-PC (1 mCi/mL) in 100 μL of PBS at 37 °C for 1.5 h. Reaction was terminated by the addition of 100 μL of chloroform/methanol (2:1, v/v). The aqueous and organic phases were extracted and dried by centrifugation under vacuum. The reaction products were resuspended in 20 μL of chloroform/methanol/acetic acid/water (23:20:5:1, v/v) and analyzed by TLC. [3H]choline was separated on a silica gel 60 HPTLC plate using chloroform/methanol/acetic acid/water (23:20:5:1, v/v) as the developing solvent. The plate was sprayed with EN3HANCE (Perkin Elmer Life and Analytical Sciences) and exposed to Hyperfilm ECL (GE Healthcare) for 1 day at −80 °C.
In vitro PLD assay for MS
For the in vitro PLD assay using MALDI-QIT-TOF/MS, 25 nmol of DPPC was rehydrated in 100 μL of PBS (pH 7.4) and was sonicated using a bath type sonicator. The final DPPC concentration was 250 μM. The immunoprecipitated PLD2-HA was incubated with DPPC in PBS at 37 °C for 3 h. Reaction was terminated by the addition of 100 μL of chloroform. The aqueous and organic phases were extracted and dried by centrifugation under vacuum. For inhibitory experiments, PLD2-HA was incubated with 100–500 nM of FIPI (diluted from a 10 mM stock concentration in DMSO).
Results
Identification of a matrix for detecting choline and PA
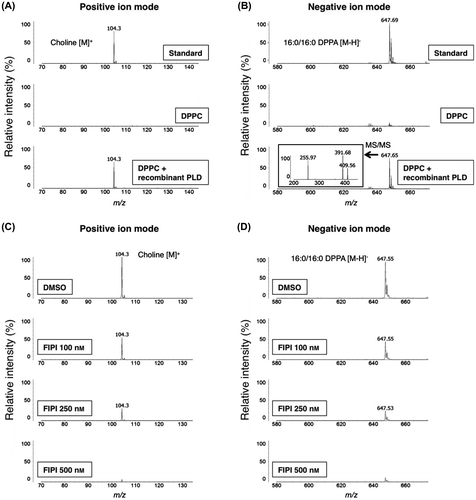
Recent advances in MALDI-MS have allowed the rapid and sensitive detection of phospholipidsCitation12); however, the choice of organic matrix is difficult because of requiring good absorptivity at the given laser wavelength, good solubility in solvents, suitable acidity and basicity, and high ionization efficiency of molecules. To determine a proper matrix that enables to detect choline and PA, both of which are products of hydrolysis of PC by PLD, we tested suitability of three matrices: DHBA, CHCA, and 9-AA. We prepared choline and PA as standards and performed the MALDI-QIT-TOF/MS analysis using the above three matrices. In positive ion mode, molecular ion of choline was observed at m/z 104.3 in all matrices (Fig. (A)). In particular, 9-AA had clearly higher intensity and lower background than DHBA and CHCA, respectively, suggesting that 9-AA is the most suitable matrix for detecting choline (Fig. (A)).
Fig. 1. MALDI-QIT-TOF/MS analysis for detecting choline and pa with different matrices.
Notes: MS spectra of choline and PA detected by MALDI-QIT-TOF/MS with DHBA (upper panel), CHCA (middle panel), and 9-AA (lower panel) as matrices in positive ion mode (A) and in negative ion mode (B) and representative MS/MS spectra [M–H]− (m/z 647.5) of standard PA (C). Peaks are indicated according to their m/z position and peaks denoted by * originated from matrix ions of CHCA.
![Fig. 1. MALDI-QIT-TOF/MS analysis for detecting choline and pa with different matrices.Notes: MS spectra of choline and PA detected by MALDI-QIT-TOF/MS with DHBA (upper panel), CHCA (middle panel), and 9-AA (lower panel) as matrices in positive ion mode (A) and in negative ion mode (B) and representative MS/MS spectra [M–H]− (m/z 647.5) of standard PA (C). Peaks are indicated according to their m/z position and peaks denoted by * originated from matrix ions of CHCA.](/cms/asset/9ba5d0e0-fbdb-4acc-9046-b8d93c209c5b/tbbb_a_910102_f0001_b.gif)
We next attempted to detect PA in negative ion mode, but neither DHBA nor CHCA produced signal intensity for PA, likely due to its relatively higher acidity (Fig. (B)). In contrast, only 9-AA exhibited a strong [M–H]− ion peak at m/z 647.5 (Fig. (B), bottom). To further validate whether this ion peak indicates PA, the MS/MS analysis was performed and proved to be an identical fragment ion of DPPA (from the left: palmitic acid [M–H]− at m/z 256.0, 16:0 lysophosphatidic acid (LPA) [M–H2O]− at m/z 391.8, 16:0 LPA [M–H]− at m/z 409.7) (Fig. (C)). Taken together, these results suggest that 9-AA matrix can be available for measuring not only choline but also PA in the MALDI-QIT-TOF/MS analysis.
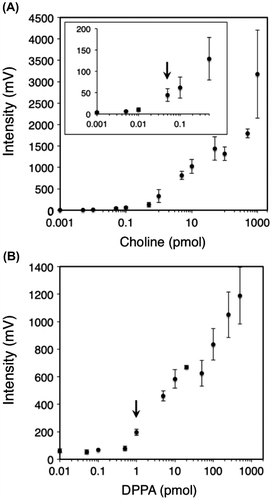
Determination of the minimum detection limits of choline and PA with 9-AA matrix
To determine the detection limits of choline and PA in 9-AA matrix, the analyzed concentration of both standards was gradually increased from 1 fmol to 1 nmol. The calibration curve for choline demonstrated that the linearity, defined as signal-to-noise ratio of 3:1, is over the range from 50 fmol (Fig. (A)-inset, arrow). On the other hand, the linearity of PA was determined over the range from 1 pmol (Fig. (B), arrow). These results indicate that at least more than 1 pmol of choline or PA is sufficiently high to be detected with 9-AA matrix in the MALDI-QIT-TOF/MS analysis.
Fig. 2. Detection limits of choline and PA with 9-AA matrix by MALDI-QIT-TOF/MS.
Notes: Standard curves for determining choline (A) and PA (B) from 1 fmol to 1 nmol with 9-AA matrix (20 mg/mL). The detection limits are defined as signal-to-noise ratio of 3:1. Each point represents the mean ± SD of triplicate measurements.
Detection of bacterial PLD-derived choline and PA from PC
Because PLDs hydrolyze the phosphodiester bond of PC, resulting in the production of choline and PA,Citation5) we tested whether choline and PA yielded by hydrolysis of PC in vitro can be detected by using 9-AA matrix. A commercially available PLD from Streptomyces sp. was used as recombinant protein, followed by incubation with DPPC, and then the reaction solutions were analyzed by the MALDI-TOF/MS in both positive and negative ion modes. We found that the reaction products’ mass spectra were identical to those of choline and PA standards only after incubation with recombinant PLD (Fig. (A) and (B)). Furthermore, MS spectra of MS/MS analysis in negative ion mode showed the same mass-to-charge ratio peaks of PA (Fig. (B)-inset). To ascertain whether the analytical peaks of choline and PA are indeed derived from the hydrolysis by PLD, we conducted an in vitro lipase-inhibition assay by using 5-fluoro-2-indolyldes-clorohalopemide (FIPI) as a PLD-specific inhibitor.Citation24) PC was incubated with recombinant PLD with an increasing amount of FIPI from 100 to 500 nM, and then the reaction products were analyzed by the MALDI-TOF/MS. As shown in Fig. (C) and (D), signal intensities of both choline and PA were deceased in a dose-dependent manner. These results suggest that the utilization of 9-AA matrix allows the concomitant detection of choline and PA as the hydrolysis products of PC.
Fig. 3. Measuring bacterial PLD activity by MALDI-QIT-TOF/MS.
Notes: MS spectra of bacteria PLD-derived products with 9-AA as matrix in positive ion mode (A) and in negative ion mode (B). Upper panel: standard choline or PA with buffers but no enzyme. Middle panel: DPPC substrate with buffers but no enzyme. Lower panel: DPPC substrate incubated with the Streptomyces sp. PLD resulting in the conversion of a portion of DPPC to the expected hydrolyzed product. The inset displays MS/MS spectrum of peak at m/z 647.65 (B). Inhibition of PC hydrolysis by Streptomyces sp. PLD was performed in the presence of FIPI (C and D).
Detection of mammalian PLD2-derived choline and PA from PC
We next investigated whether our method could be used to examine the enzymatic activity of mouse PLD by measuring choline and PA in the reaction solution. As mammalian PLD2 exhibits high basal constitutive activity when assayed in vitro,Citation8) we immunopurified the overexpressed mouse PLD2 proteins from HEK293T cells. Furthermore, to exclude the possibility that choline and PA are derived solely by contamination from cell extracts, we prepared a catalytically inactive mutant containing a lysine to arginine substitution within the HKD motif (K758R) as a negative control.Citation25) Similar expression levels of PLD2-HA proteins were observed between wild-type and the K758R mutant (Fig. (A)). We first attempted to confirm the enzymatic activities of the immunoprecipitated PLD2 by the TLC analysis. After incubation with purified PLD2 and [choline-methyl-3H]dipalmitoyl-PC, chloroform/methanol (2:1, v/v) was added to stop the reaction and then separated in the aqueous and organic phases. As expected, wild-type, but not the K758R mutant of PLD2, produced [3H]choline as water-soluble product (Fig. (B)), indicating that the purified PLD2 from HEK 293T is able to catalyze the hydrolysis of PC in vitro.
Fig. 4. Measuring mammalian PLD2-derived products from PC.
Notes: HEK293T cells were transfected with plasmids expressing control vector, mouse PLD2 wild-type or catalytically inactive K758R mutant, as indicated. PLD2-HA immunoprecipitates were immunoblotted with antibody against HA (A). Control vector, mouse PLD2 wild-type or K758R mutant purified proteins were incubated with 3H-labeled DPPC, and then the water-soluble products were analyzed by TLC (B). TLC was carried out as described in Materials and Methods. The standard products were [3H]PC and [3H]choline chloride. MS spectra of control vector, mouse PLD2 wild-type or K758R mutant-derived products with 9-AA as matrix in positive ion mode (C) and in negative ion mode (D) as indicated. Upper panel: standard choline or PA with buffers but no enzyme. The inset displays MS/MS spectrum of peak at m/z 647.57.
![Fig. 4. Measuring mammalian PLD2-derived products from PC.Notes: HEK293T cells were transfected with plasmids expressing control vector, mouse PLD2 wild-type or catalytically inactive K758R mutant, as indicated. PLD2-HA immunoprecipitates were immunoblotted with antibody against HA (A). Control vector, mouse PLD2 wild-type or K758R mutant purified proteins were incubated with 3H-labeled DPPC, and then the water-soluble products were analyzed by TLC (B). TLC was carried out as described in Materials and Methods. The standard products were [3H]PC and [3H]choline chloride. MS spectra of control vector, mouse PLD2 wild-type or K758R mutant-derived products with 9-AA as matrix in positive ion mode (C) and in negative ion mode (D) as indicated. Upper panel: standard choline or PA with buffers but no enzyme. The inset displays MS/MS spectrum of peak at m/z 647.57.](/cms/asset/0842b41a-1c45-4444-96e9-de237b85ad2d/tbbb_a_910102_f0004_b.gif)
Next, we assessed whether the 9-AA-based MALDI-MS method directly detects the PC-derived choline and PA catalyzed by immunopurified PLD2. In both positive and negative ion modes, the mass spectra of choline and PA were readily observed in the reaction products only when incubated with wild-type PLD2 (Fig. (C) and (D)). Moreover, MS spectra of MS/MS analysis in negative ion mode showed the same mass-to-charge ratio of PA (Fig. (D)-inset). Finally, to examine the effect of inhibition on PLD activity, FIPI was added to the same in vitro PLD2 assay as in Fig. (C) and (D). As shown in Fig. S1A and S1B, FIPI decreased signal intensities of both choline and PA in a dose-dependent manner (Supplemental Fig. 1; see Biosci. Biotechnol. Biochem Website). These results show that our method can be used to evaluate PLD2 activity by directly detecting choline and PA as reaction products.
Discussion
Current progress of matrices development has enabled us to analyze various cellular phospholipids using the MALDI-TOF/MS spectrometry.Citation12,15,26) Here, we report a simple, sensitive, and rapid method for detecting PA as well as choline by using the 9-AA matrix. Furthermore, we found that this method can be available for the simultaneous detection of choline and PA followed by PLD-mediated hydrolysis of PC. In contrast to the previous methods whereby either choline or PA is measured with the Amplex Red system,Citation10,11) our present method can detect both choline and PA directly, using MALDI-QIT-TOF/MS, for evaluating PLD activity in vitro.
Among three matrices: DHBA, CHCA, and 9-AA, our initial investigation revealed that standard choline can be detected most clearly in 9-AA without background interference, especially from m/z 100 to 130 (Fig. (A)). In addition, we have confirmed that 9-AA alone has no MS signal in the vicinity of m/z 104 (data not shown). A plausible explanation for the chemical properties of 9-AA may be that it functions as a competitor for protons and thereby reduces the matrix background in the MS spectrum.Citation21,27) Since 9-AA has been shown to yield low background and good sensitivity for selected low molecular weight due in part to the presence of one basic amine moiety on a conjugated ring system, the 9-AA-based MALDI-MS method appears to be one of the powerful techniques for detecting choline in positive ion mode.
Previous studies have shown that PA is quite difficult to be detected in negative ion MS spectra using DHBA matrix, even when applying higher concentrations (up to 3 mg/mL).Citation28) In contrast, our present method using 9-AA could detect PA ion (m/z 647) from 1 pmol in negative ion mode (Fig. (B)). Given that PA bears two negative chargesCitation28,29) and the addition of one positive ion is necessary for the detection of PA as a singly charged ion, it appears that 9-AA can transfer proton to the PA molecule, which in turn forms a single negative charge of PA, [M–H]−. Actually, we demonstrated that 9-AA exhibits low background and high sensitivity for selected PA fragment ions, especially from m/z 200 to 450 under CID MS/MS conditions (Fig. (C)). Thus, our data provide first evidence that PA can be successfully and efficiently detected by using 9-AA matrix in MALDI-QIT-TOF/MS spectrometry.
In the study, we showed that this method was able to directly detect PA catalyzed by PLD using MALDI-QIT-TOF/MS (Figs. (B) and (D)). In previous methods using the Amplex Red system, whereby either measurements of choline or PA is included in the two- or three-step processes with enzymes for generating hydrogen peroxide.Citation10,11) In contrast, the major advantage of our method is to detect PLD-derived products directly in a single step, which makes it useful, for example, to assess the ability of inhibitor or mutation experiments of PLD, an important topic of interest in the phospholipid regulation.
Unlike PLD2 with constitutive activity, mammalian PLD1 has been shown to require intracellular regulators, such as phosphatidylinositol-4,5-bisphosphate (PI4,5P2), protein kinase C (PKC), ADP-ribosylation factor (ARF), and Rho family GTPases, to enhance the basal activity.Citation5,8,30) However, the cellular substrates and optimum conditions of additional PLD members still remain elusive and further investigation with new analytical techniques are needed to accurately evaluate the activity of PLD. In this point, our single-step method could be utilized only by adding the candidates into the reaction solution of PC hydrolysis. Therefore, such an analysis for PLD-derived products using our method can give rise to valuable information about the enzymatic activity of PLDs as in vitro studies.
Recent studies reported that choline and PA were detected using the method of liquid chromatography–mass spectrometry (LC–MS).Citation31,32) It has an impact on quantitative analysis of these molecules and can be online coupled with a chromatographic column to separate them from impurities inhibiting the ionization of target molecules. On the other hand, MALDI-QIT-TOF/MS method enables us to analyze choline and PA directly without any separation process prior to measurement of molecular mass. Furthermore, the method allows to optimize analytical conditions (ex. sensitivity, signal/noise ratio, etc.) through selecting matrices. In the present study, we succeeded in enhancing sensitivity by adopting 9-AA as a matrix, rather than DHBA and/or CHCA, and detecting choline and PA, not only in standard molecules, but also in PLD-derived products from PC. This method will be useful for future studies on biological functions of PLD.
Supplemental material
The supplemental material for this paper is available at http://dx.doi.10.1080/09168451.2014.910102.
Supplemental Fig.2 caption
Download MS Word (13 KB)Supplemental Fig.1
Download PDF (182.8 KB)Supplemental Fig.2
Download MS Power Point (204 KB)Acknowledgments
We thank Dr. Yasunori Kanaho (University of Tsukuba, Japan) and Dr. Michael A. Frohman (Stony Brook University, USA) for plasmids of mouse PLD2 wild-type and K758R mutant. This work has been supported by the Grant-in-Aid for Scientific Research (A) [grant number 22248040].
References
- Kent C. Annu. Rev. Biochem. 1995;64:315–343.10.1146/annurev.bi.64.070195.001531
- Ridgway ND. Crit. Rev. Biochem. Mol. Biol. 2013;48:20–38.10.3109/10409238.2012.735643
- Van Meer G, Voelker DR, Feigenson GW. Nat. Rev. Mol. Cell Biol. 2008;9:112–124.10.1038/nrm2330
- Cole LK, Vance JE, Vance DE. Biochim. Biophys. Acta 2012;1821:754–761.10.1016/j.bbalip.2011.09.009
- Selvy PE, Lavieri RR, Lindsley CW, Brown HA. Chem. Rev. 2011;111:6064–6119.10.1021/cr200296t
- Wang X, Devaiah SP, Zhang W, Welti R. Prog. Lipid Res. 2006;45:250–278.10.1016/j.plipres.2006.01.005
- Jenkins GM, Frohman MA. Cell. Mol. Life Sci. 2005;62:2305–2316.10.1007/s00018-005-5195-z
- Colley WC, Sung TC, Roll R, Jenco J, Hammond SM, Altshuller Y, Bar-Sagi D, Morris AJ, Frohman MA. Curr. Biol. 1997;7:191–201.10.1016/S0960-9822(97)70090-3
- Morris AJ, Frohman MA, Engebrecht J. Anal. Biochem. 1997;252:1–9.10.1006/abio.1997.2299
- Balcerzak M, Pikula S, Buchet R. FEBS Lett. 2006;580:5676–5680.10.1016/j.febslet.2006.09.018
- Morita SY, Ueda K, Kitagawa S. J. Lipid Res. 2009;50:1945–1952.10.1194/jlr.D900014-JLR200
- Schiller J, Süß R, Arnhold J, Fuchs B, Lessig J, Müller M, Petković M, Spalteholz H, Zschörnig O, and Arnold K. Prog. Lipid Res. 2004;43:449–488.10.1016/j.plipres.2004.08.001
- March RE. Mass Spectrom. Rev. 2009;28:961–989.10.1002/mas.v28:6
- Beavis RC, Chait BT. Anal. Chem. 1990;62:1836–1840.10.1021/ac00216a020
- Fuchs B, Schiller J. Eur. J. Lipid Sci. Technol. 2009;111:83–98.10.1002/ejlt.v111:1
- Beavis RC, Chaudhary T, Chait BT. Org. Mass Spectrom. 1992;27:156–158.10.1002/(ISSN)1096-9888
- Strupat K, Karas M, Hillenkamp F. Int. J. Mass Spectrom. Ion Processes. 1991;111:89–102.10.1016/0168-1176(91)85050-V
- Angelini R, Babudri F, Lobasso S, Corcelli A. J. Lipid Res. 2010;51:2818–2825.10.1194/jlr.D007328
- Schiller J, Süß R, Fuchs B, Müller M, Petković M, Zschörnig O, Waschipky H. Eur. Biophys. J. 2007;36:517–527.10.1007/s00249-006-0090-6
- Guo Z, He L. Anal. Bioanal.Chem. 2007;387:1939–1944.10.1007/s00216-006-1100-3
- Sun G, Yang K, Zhao Z, Guan S, Han X, Gross RW. Anal. Chem. 2008;80:7576–7585.10.1021/ac801200w
- Brown HA, Gutowski S, Moomaw CR, Slaughter C, Sternwels PC. Cell 1993;75:1137–1144.10.1016/0092-8674(93)90323-I
- Lopez I, Arnold RS, Lambeth JD. J. Biol. Chem. 1998;273:12846–12852.10.1074/jbc.273.21.12846
- Su W, Yeku O, Olepu S, Genna A, Park JS, Ren H, Du G, Gelb MH, Morris AJ, Frohman MA. Mol. Pharmacol. 2009;75:437–446.10.1124/mol.108.053298
- Sung TC, Roper RL, Zhang Y, Rudge SA, Temel R, Hammond SM, Morris AJ, Moss B, Engebrecht J, Frohman MA. EMBO J. 1997;16:4519–4530.10.1093/emboj/16.15.4519
- Loizides-Mangold U. FEBS J. 2013;280:2817–2829.10.1111/febs.2013.280.issue-12
- Fuchs B, Bischoff A, Süß R, Teuber K, Schürenberg M, Suckau D, Schiller J. Anal. Bioanal.Chem. 2009;395:2479–2487.10.1007/s00216-009-3032-1
- Petković M, Schiller J, Müller M, Benard S, Reichl S, Arnold K, Arnhold J. Anal. Biochem. 2001;289:202–216.10.1006/abio.2000.4926
- Petković M, Schiller J, Müller M, Suss R, Arnold K, Arnhold J. Z. Naturforsch. B: Chem. Sci. 2009;64:331–334.
- Henage LG, Exton JH, Brown HA. J. Biol. Chem. 2006;281:3408–3417.10.1074/jbc.M508800200
- Holm PI, Ueland PM, Kvalheim G, Lien EA. Clin. Chem. 2003;49:286–294.10.1373/49.2.286
- Mizuno S, Sakai H, Saito M, Kado S, Sakane F. FEBS Open Bio. 2012;2:267–272.10.1016/j.fob.2012.08.006
