Abstract
Impaired protein folding in the endoplasmic reticulum (ER) evokes the unfolded protein response (UPR), which is triggered in budding yeast, Saccharomyces cerevisiae, by the ER-located transmembrane protein Ire1. Here, we report that ethanol stress damages protein folding in the ER, causing activation of Ire1 in yeast cells. The UPR likely contributes to the ethanol tolerance of yeast cells.
The endoplasmic reticulum (ER) is a cellular compartment, where secretory proteins are folded and membrane lipids are synthesized. ER proteins, including the ER-located molecular chaperone BiP, are transcriptionally induced under ER-stress conditions in which ER functions are impaired. In Saccharomyces cerevisiae (hereafter termed yeast) cells, this cellular response is triggered by the ER-located type-I transmembrane RNase Ire1. During conditions of ER stress, Ire1 self-associates to form large oligomers and splices the HAC1-gene transcript (HAC1u) to yield translatable mRNA (HAC1i) that is translated into a transcription-factor protein.
Since this cellular response is evoked by the accumulation of unfolded proteins in the ER, it has been termed the unfolded protein response (UPR). The luminal domain of Ire1 captures unfolded proteins accumulated in the ER, leading to the oligomerization and activation of Ire1.Citation1) In laboratory experiments, such stress stimuli are often induced by culturing cells with the disulfide-bond cleaving reagent dithiothreitol (DTT). Ire1 is also activated by the disturbance of membrane lipid homeostasis. For example, Ire1 is activated by depletion of the membrane lipid component inositol through an unknown mechanism.Citation2) Thus, Ire1 is activated, and the UPR is induced by dual and distinct processes.
The ethanol tolerance of yeast cells is an industrially important matter concerning high productivity during ethanol fermentation. We previously reported that the UPR is transiently evoked when cells were exposed to 8% ethanol.Citation3) Brown et al.Citation4) recently reported that the UPR is induced during the fermentation process in bioethanol fermentation strains that are highly tolerant to ethanol, but not in the laboratory strain S288c, suggesting UPR involvement in ethanol tolerance. However, to date, no further evidence showing a contribution of the UPR to ethanol tolerance has been presented. Moreover, the mechanism by which ethanol stress leads to evocation of the UPR remains unclear.
In this study, laboratory yeast strains derived from the ire1Δ haploid strain KMY1015Citation3) were cultured at 30 °C in standard synthetic dextrose (SD) mediumCitation1) and sequentially stressed by 8% ethanol for 4 h and then 16% ethanol. The reason for the stepwise incremental increase of ethanol concentration in the medium was to adapt the yeast cells to potent ethanol stress. It should be also noted that ethanol concentration gradually increases during industrial ethanol fermentation. In the experiment shown in Fig. (A), the HAC1-mRNA species were amplified from total RNA samples by RT-PCR as described previously.Citation2) The efficiency of mRNA splicing is quantitatively presented in Fig. (B). We then found that stressing cells with 16% ethanol caused sustained HAC1-mRNA splicing, unlike the effect of stressing with only 8% ethanol, where Ire1 was transiently activated (peak activation was seen 30 min after the onset of ethanol stressCitation3)).
Fig. 1. Ire1 activation upon ethanol stress.
Notes: (A) IRE1+ cells (the ire1Δ haploid strain transformed with a centromeric IRE1 plasmid, pRS313-IRE1Citation3)) were exponentially grown in SD medium (lane 1) and stressed by addition of ethanol (8% (v/v) final concentration) to the culture, which was incubated for another 4 h (lane 2). Ethanol was then added again to give a final concentration of 16% (v/v), and cells were further incubated for the indicated durations (lanes 3–8). Total RNA samples were subjected to RT-PCR using the HAC1-specific PCR primer set,Citation2) and the products were run on 2% agarose. (B) The same experiment shown in panel A was performed using three independent clones. The HAC1-mRNA splicing efficiency was calculated and is presented as the mean plus standard deviation. (C) and (D) Ire1-GFP was expressed in the ire1Δ strain, and its GFP fluorescent images were pictured as described previously.Citation6) (E) IRE1+ cells and ΔIII-Ire1 cells (the ire1Δ strain transformed with a ΔIII-mutant version of pRS313-IRE1Citation1)) were stressed by the indicated stimuli. Splicing of the HAC1 mRNA was monitored as described for the experiments shown in panels A and B.
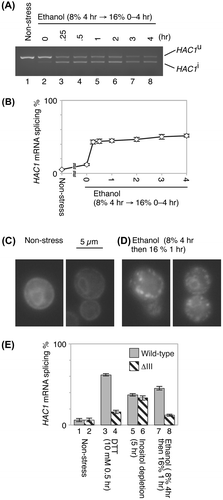
As mentioned above, activation of Ire1 is accompanied by its high-order oligomerization,Citation1,5) which can be visualized as punctate fluorescent spots when Ire1 is tagged with GFP (Ire1-GFP).Citation5,6) Similar to the observations in previous reports,Citation5,6) the Ire1-GFP fluorescence exhibited double ring-like ER patterns that are typical for non-stressed cells (Fig. (C)), indicating that it was diffusively distributed over the ER. However, as shown in Fig. (D), punctate spots of Ire1-GFP were observed in cells stressed by 16% ethanol.
These observations indicate that potent ethanol stress induces the UPR in canonical fashion, namely the oligomerization and activation of Ire1, followed by splicing of the HAC1 mRNA. It should also be noted that induction of the UPR by ethanol stress is an issue that goes beyond the industrial ethanol fermentation strains.
The ΔIII mutant of yeast Ire1 carries a partial deletion of the luminal domain and is impaired in capturing unfolded proteins.Citation1,2) We previously reported that the ΔIII mutation compromises UPR induced by DTT exposure, which causes accumulation of unfolded proteins in the ER, but not by inositol depletion, which causes membrane lipid aberrancy.Citation2) This observation is reproduced in Fig. (E) (compare column 4–3 and column 6–5). We then examined ethanol stress and, as shown in Fig. (E) (columns 7 and 8), found that Ire1 activation was compromised in the ΔIII mutant. This finding strongly suggests that ethanol stress induces the UPR through accumulation of unfolded proteins in the ER.
We then investigated whether ethanol stress actually impairs protein folding in the ER. Kar2, the yeast BiP orthologue, is known to be often incorporated into aggregates of unfolded proteins accumulated in the ER. In our previous studies, we lysed cells in the presence of the mild detergent Triton X-100 and subsequently fractionated them by centrifugation. We then showed that Kar2 was incorporated into the pellet fractions when cells were stressed by DTT or by heterologous expression of a model-unfolded protein, but not by inositol depletion.Citation2,7) By using the same methodology, we showed in this study that stressing cells with 16% ethanol caused incorporation of Kar2 into the pellet fraction (Fig. (A)).
Fig. 2. BiP aggregation upon ethanol stress.
Notes: (A) The IRE1+ cells remained unstressed or were exposed to ethanol stress. Total cell lysates were then fractionated by centrifugation at 8000 × g for 20 min. The supernatants (equivalent to 0.1 OD600 cells) and pellets (equivalent to 1.0 OD600 cells) were analyzed by anti-BiP western blotting.Citation2) (B) and (C) Cells were immunofluorescently stained with anti-Kar2 antiserum and pictured as described previously.Citation1) (D) The anti-Kar2 immunofluorescence-stained images were used to count cells with “punctate Kar2 distribution”, in which the nuclear ER was not observed as a closed ring. More than 100 anti-Kar2 immunofluorescently stained cells per specimen were assessed. Data are presented as the means plus standard deviations from multiple determinations. (E) kar2-1IRE1+ cells (KMY81Citation7)) were stressed by ethanol at the semi-permissive temperature of 30 °C and analyzed as done in panels B and C. (F) and (G) IRE1+ cells transformed with the eroGFP expression plasmid pPM28Citation8) were observed under a Deltavision microscope (Applied Precision).
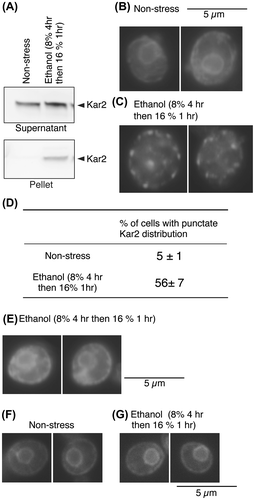
We hypothesized that if protein aggregates form large punctate structures in the ER, then cellular distribution of Kar2 might be visualized as punctate spots. Indeed, Kar2 exhibited a normal ER-like distribution pattern in non-stressed cells (Fig. (B)), while ethanol stress caused a punctate distribution of Kar2 (Fig. (C)). This observation is quantitatively presented in Fig. (D). It should be noted that punctate distribution of Kar2 was not observed when it carried the kar2-1 mutation (Fig. (E)), which impairs association between unfolded proteins and Kar2.Citation7) This observation indicates that Kar2 aggregates actually through its association with unfolded proteins. The fluorescent images of the ER-targeted GFP (eroGFP)Citation8) indicate that the broad outline of ER morphology (the double ring) was not disturbed by ethanol stress (Fig. (F) and (G)).
Finally, we evaluated whether or not the UPR actually contributes to ethanol tolerance. To this end, cells carrying or not carrying the IRE1 gene were stressed by ethanol (8%, 4 h; then 16%, 24 h) before being plated onto normal SD agar plates. As quantitatively presented in Fig. , we observed that potent ethanol stress reduced the viability of ire1Δ cells more than that of IRE1+ cells. A similar observation was obtained when we compared viability of hac1Δ cells to that of wild-type cells.
Fig. 3. Reduction of ethanol tolerance by the ire1Δ or the hac1Δ mutation.
Notes: (A) IRE1+ cells and ire1Δ cells (the ire1Δ strain transformed with pRS313-IRE1 or the empty vector pRS313) were treated with ethanol (“8% 4 h only” or “8% ethanol then 16% 24 h”) and plated onto non-stressing SD agar plates. After incubation for 3 days, colony numbers on the agar plates were counted to calculate “survival %” using the formula 100X [colony number from “8% ethanol then 16% 24 h” sample]/[colony number from “8% 4 h only” sample]. Data are presented as the means and standard deviations from 3 independent transformants. (B) The same experiments were performed using wild-type strain KMY1005 and hac1Δ strain KMY1045.Citation1) Data are presented as the means and standard deviations from 3 independent determinations.
![Fig. 3. Reduction of ethanol tolerance by the ire1Δ or the hac1Δ mutation.Notes: (A) IRE1+ cells and ire1Δ cells (the ire1Δ strain transformed with pRS313-IRE1 or the empty vector pRS313) were treated with ethanol (“8% 4 h only” or “8% ethanol then 16% 24 h”) and plated onto non-stressing SD agar plates. After incubation for 3 days, colony numbers on the agar plates were counted to calculate “survival %” using the formula 100X [colony number from “8% ethanol then 16% 24 h” sample]/[colony number from “8% 4 h only” sample]. Data are presented as the means and standard deviations from 3 independent transformants. (B) The same experiments were performed using wild-type strain KMY1005 and hac1Δ strain KMY1045.Citation1) Data are presented as the means and standard deviations from 3 independent determinations.](/cms/asset/0394250e-26f9-440f-b910-1da41702840a/tbbb_a_921561_f0003_b.gif)
Ethanol is known to work as a protein denaturant. According to Trotter et al.,Citation9) ethanol stress denatures nuclear and cytosolic proteins and triggers the heat shock response in yeast cells. Here, we propose that ethanol stress in yeast cells also damages ER protein folding, forms protein aggregates in the ER, and induces the UPR, which contributes to ethanol tolerance. Although ethanol is known to directly affect membrane properties, this is unlikely to be the main cause of the ethanol-induced UPR. Considering the strong UPR induction in ethanol-tolerant industrial strains reported by Brown et al.Citation4) along with our results, artificial manipulation of the UPR pathway might be a useful strategy to breed industrially valuable yeast strains.
Funding
This work is supported by MEXT/JSPS KAKENHI [grant number 22657030] and [grant number 24370081] to Y.K.
Notes
Abbreviations: DTT, dithiothreitol; ER, endoplasmic reticulum; GFP, green fluorescent protein; RT-PCR, reverse transcription PCR; SD, synthetic dextrose; UPR, unfolded protein response.
References
- Kimata Y, Ishiwata-Kimata Y, Ito T, Hirata A, Suzuki T, Oikawa D, Takeuchi M, Kohno K. Two regulatory steps of ER-stress sensor Ire1 involving its cluster formation and interaction with unfolded proteins. J. Cell Biol. 2007;179:75–86.10.1083/jcb.200704166
- Promlek T, Ishiwata-Kimata Y, Shido M, Sakuramoto M, Kohno K, Kimata Y. Membrane aberrancy and unfolded proteins activate the endoplasmic reticulum stress sensor Ire1 in different ways. Mol. Biol. Cell. 2011;22:3520–3532.10.1091/mbc.E11-04-0295
- Kimata Y, Oikawa D, Shimizu Y, Ishiwata-Kimata Y, Kohno K. A role for BiP as an adjustor for the endoplasmic reticulum stress-sensing protein Ire1. J. Cell Biol. 2004;167:445–456.10.1083/jcb.200405153
- Brown NA, de Castro PA, de Castro Pimentel Figueiredo B, Savoldi M, Buckeridge MS, Lopes ML, de Lima Paullilo SC, Borges EP, Amorim HV, Goldman MH, Bonatto D, Malavazi I, Goldman GH. Transcriptional profiling of Brazilian Saccharomyces cerevisiae strains selected for semi-continuous fermentation of sugarcane must. FEMS Yeast Res. 2013;13:277–290.10.1111/fyr.2013.13.issue-3
- Aragón T, van Anken E, Pincus D, Serafimova IM, Korennykh AV, Rubio CA, Walter P. Messenger RNA targeting to endoplasmic reticulum stress signalling sites. Nature. 2009;457:736–740.10.1038/nature07641
- Ishiwata-Kimata Y, Yamamoto YH, Takizawa K, Kohno K, Kimata Y. F-actin and a type-II myosin are required for efficient clustering of the ER stress sensor Ire1. Cell Struct. Funct. 2013;38:135–143.10.1247/csf.12033
- Kimata Y, Kimata YI, Shimizu Y, Abe H, Farcasanu IC, Takeuchi M, Rose MD, Kohno K. Genetic evidence for a role of BiP/Kar2 that regulates Ire1 in response to accumulation of unfolded proteins. Mol. Biol. Cell. 2003;14:2559–2569.10.1091/mbc.E02-11-0708
- Merksamer PI, Trusina A, Papa FR. Real-time redox measurements during endoplasmic reticulum stress reveal interlinked protein folding functions. Cell. 2008;135:933–947.10.1016/j.cell.2008.10.011
- Trotter EW, Kao CM, Berenfeld L, Botstein D, Petsko GA, Gray JV. Misfolded proteins are competent to mediate a subset of the responses to heat shock in Saccharomyces cerevisiae. J. Biol. Chem. 2002;277:44817–44825.10.1074/jbc.M204686200
