Abstract
Cells of Lactobacilli co-aggregated with Escherichia coli K-12 cells to form co-aggregates under mixed-culture conditions at 37 °C for 24 h. Co-aggregation was inhibited by sodium dodecyl sulfate but not by protease. E. coli deletion mutants of fimbriae formation and lipopolysaccharide (LPS) formation did not co-aggregate with Lactobacilli. These results showed that fimbriae and LPS are necessary for co-aggregation between Lactobacilli and E. coli.
Lactobacilli are usually found in various environments including human intestine, and they are considered to contribute to human health by controlling the intestinal microflora. However, precise functions of the probiotic Lactobacilli in controlling microflora of the intestine have not been understood well.Citation1–4)
Lactobacilli produce lactic acid, reducing the environmental pH,Citation5) and therefore, they can produce acidic environmental niches, such as the oral cavity.Citation6) Some studies showed that oral administration of Lactobacilli increased the levels of Lactobacilli in human feces and also decreased the numbers of fecal Escherichia coli and Anaerobic cocci.Citation7,8) These studies indicated that lower pH environment brought about by lactic acid could decrease the other bacteria such as Enterobacteriaceae.
In the last two decades, adhesive nature of Lactobacilli in human intestine has been well studied.Citation1,9–12) It was showed that some probiotic Lactobacilli could bind to human epithelial cells, and they also could co-aggregate with the commensal bacterium Enterococcus faecium.Citation10) Several Lactobacilli including isolates from human body have been reported to co-aggregate with E. coli.Citation1,10) These studies showed that co-aggregation would be an important factor for Lactobacilli in controlling microflora in human body.
Co-aggregation is cell-to-cell adhesion between different types of cells, and that is observed in various environments, including the human body.Citation13–16) Co-aggregation is induced by non-specific physicochemical or specific biological interactions.Citation17–19)
Here, we investigated that the co-aggregative properties and mechanisms of various Lactobacilli cells with E. coli cells. This study would provide us new information on the function of Lactobacilli in our intestine.
E. coli K-12 MG1655 (wild type; F−, lambda−, rph-1) was used in this study. Some E. coli deletion strains, ΔfliC (JW1908), Δfim (JW4277), ΔcsgA (JW1024), and ΔrfaC (JW3596), of KEIO collection including parent wild-type strain E. coli K-12 BW25113 were obtained from National Institute of Genetics (Shizuoka, JAPAN).Citation20) Lactobacilli strains and some other lactic acid bacteria (LAB) strains used in this study were described in Table . E. coli was cultured in Luria-Bertani (LB) medium (Becton, Dickinson, Sparks, MD) at 37 °C for 24 h with shaking at 120 rpm. LAB were cultured statically in De Man, Rogosa, and Sharpe broth (MRS; Oxoid, Hampshire, England) at 37 °C for 24 h.
Table 1. Co-aggregation between E. coli MG1655 and various LAB.
Unless otherwise stated, all the reagents used in this study were of reagent grade and were purchased from Kanto Chemical (Tokyo), Wako Pure Chemical Industries (Osaka, Japan), or Sigma-Aldrich (St. Louis, MO).
For assaying co-aggregation, the 2 mL per Petri dish (or 10 μL per well of a 24-multiwell plate) of E. coli 24 h culture was mixed with 1 mL per dish (or 50 μL per well) of LAB 24 h culture. Then, the mixtures were incubated at 37 °C for 24 h, and co-aggregates were used for further analyses.
To assess the effects of various inhibitors on co-aggregation of E. coli MG1655 with L. casei subsp. rhamnosus NBRC3831, 1 mg/mL proteinase K and pepsin; 1 or 3%(v/v) of methanol and butanol; 1 or 3%(w/v) Tween 20, 40, 60, 80; and 50 or 100 mM ethylenediaminetetraacetic acid (EDTA) and sodium dodecyl sulfate (SDS) were used. Temperature during co-aggregation was changed from 28 to 44 °C at intervals of 2 °C. pH of the mixtures was changed from 3.0 to 10.0 at 0.2 intervals.
Co-aggregation between E. coli and various Lactobacillus species was investigated. As shown in Table , E. coli co-aggregated with all 12 LAB tested. All tested LAB did not form aggregates without E. coli cells. For further analyses, we used L. casei subsp. rhamnosus NBRC3831 for its higher co-aggregation activity.
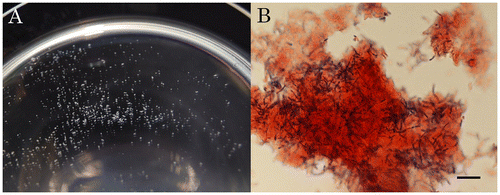
Large co-aggregates of E. coli and L. casei were observed by microscopy (Fig. (A)). Diameters of the visible co-aggregates ranged between 500 and 1,000 μm (Fig. (A)). Gram-staining revealed that the co-aggregates consisted of cells of E. coli and L. casei (Fig. (B). L. casei cells (black rod) appeared to be embedded in E. coli cells (light gray field) (Fig. (B)).
Fig. 1 Co-aggregation between E. coli MG1655 and L. casei NBRC3831 on a Polystyrene Dish.
Note: Unstained co-aggregates (A) and Gram-stained co-aggregates (B) are shown. Scale bar, 10 μm.
The formation of co-aggregates between E. coli and L. casei was found to be resistant to treatment with proteinase K and pepsin; methanol; butanol; Tween 20, 40, 60, and 80; and EDTA. However, it was inhibited by the addition of 50 mM SDS. These results showed that the mediator would be resistant to protease and surfactants except SDS, and that co-aggregation did not request divalent cation.
The effects of pH on the formation of co-aggregates were investigated between pH 3.0 and 10.0 at intervals of 0.2, and the co-aggregates were formed between pH 3.8 and 4.6. This result showed that acidic environment generally brought about by lactic acid would be necessary for the co-aggregation.
To identify the surface factors of E. coli necessary for co-aggregation with L. casei, the co-aggregative activities of E. coli deletion mutants lacking the genes that relate to the outer membrane structures such as flagella, curli fiber, fimbriae, and lipopolysaccharide (LPS) recognized as adhesin for various surfaces were investigated. As shown in Table , E. coli mutants that lacked fimA, gene coding the main component of fimbriae, and rfaC, gene coding forming basal core chain of LPS, lost the ability to form co-aggregates. On the other hand, E. coli mutants that lacked fliC, gene coding the main component of flagella, and csgA, gene coding the main component of curli, had the ability to form co-aggregates. These results indicated that both fimbriae and LPS are necessary for the co-aggregation of E. coli and L. casei. It has been reported that mutations in the core chain synthesis of LPS in E. coli lead to the deficiency in fimbriae biogenesis.Citation21) Therefore, ΔrfaC deleted in LPS synthesis as well as ΔfimA would lose fimbriae. Thus, fimbriae would be a main surface factor necessary for the co-aggregation.
Table 2. The co-aggregation ability of E. coli mutants lacking various outer membrane structures.
E. coli fimbriae are composed of protein subunits termed fimbrins,Citation22) whereas the fimbriae-mediated co-aggregates in this study were resistant to proteases, which is consistent with the report of the high resistance of the recombinant E. coli fimbriae to pepsinCitation23). The inhibitory effect of SDS on the co-aggregation suggested that electrostatic interaction would be also necessary for the co-aggregation. As described above, low pH was necessary for the co-aggregation, and the structure of the fimbrial protein in neutral pH would not be desirable for the co-aggregation with L. casei.
Acknowledgments
We would like to thank the National Institute of Genetics (Shizuoka) and National BioResource Project (NIG, Japan): E. coli for giving us the E. coli strains used in this study.
References
- Boris S, Suarez JE, Barbes C. Characterization of the aggregation promoting factor from Lactobacillus gasseri, vaginal isolate. J. Appl. Microbiol. 1997;83:413–420.
- Lebeer S, Vanderleyden J, De Keersmaecker C. Gene and molecules of Lactobacilli supporting probiotic action. Microbiol. Mol. Biol. Rev. 2008;72:728–764.
- Šušković J, Kos B, Goreta J, Matošić S. Role of lactic acid bacteria and bifidobacteria in synbiotic effect. Food Tech. Biotech. 2001;39:227–235.
- Tannock GW. A fresh look at the intestinal microflora. Wymondham: Horizon Scientific Press; 1999. Probiotics a critical review; p. 5–14.
- Antikainen J, Kupannen V, Laehteenmaeki K, Korhonen TK. pH-Dependent association of enolase and glyceraldehyde-3-phosphate dehydrogenase of Lactobacillus crispatus with the cell wall and lipoteichoic acids. J. Bacteriol. 2007;189:4539–4543.
- Chhour KL, Nadkarni MA, Byun R, Martin FE, Jacques NA, Hunter N. Molecular analysis of microbial diversity in advanced caries. J. Clin. Microbiol. 2005;43:843–849.
- Lidbeck A, Gustafsson JA, Nord CE. Impact of Lactobacillus acidophilus supplements on the human oropharyngeal and intestinal microflora. Scand. J. Infect. Dis. 1987;19:531–537.
- Saxelin M, Elo S, Salminen S, Vapaatalo H. Dose response colonisation of faeces after oral administration of Lactobacillus casei strain GG. Microb. Ecol. Health Dis. 1991;4:209–214.
- Alander M, Korpela R, Saxelin M, Vilpponen-Salmela T, Matilla-Sandholm T, Wright A. Recovery of Lactobacillus rhamnosus GG from human colonic biopsies. Lett. Appl. Microbiol. 1997;24:361–364.
- Kos B, Suskovic S, Simpraga M, Frece J, Matosic S. Adhesion and aggregation ability of probiotic strain Lactobacillus acidophilus M92. J. Appl. Microbiol. 2003;94:981–987.
- Pedersen K, Tannock GW. Colonization of the porcine gastrointestinal tract by lactobacilli. Appl. Environ. Microbiol. 1989;55:279–283.
- Schachtsiek M, Hammes WP, Hertel C. Characterization of Lactobacillus coryniformis DSM 2001T surface protein Cpf mediating coaggregation with and aggregation among pathogens. Appl. Environ. Microbiol. 2004;70:7078–7085.
- Gibbons RJ, Nygaard M. Inter-bacterial aggregation of plaque bacteria. Arch. Oral. Biol. 1970;15:1397–1400.
- Jin L, Tao L, Pavlova SI, So JS, Kiwanuka N, Namukwaya Z, Saberbein BA, Wawer M. Species diversity and relative abundance of vaginal lactic acid bacteria from women in Uganda and Korea. J. Appl. Microbiol. 2007;102:1107–1115.
- Kolenbrander PE. Coaggregation of human oral bacteria: potential role in the accretion of dental plaque. J. Appl. Bacteriol. 1993;74 (Suppl.):79S–86S.
- Reid G, McGroarty JA, Angotti R, Cook RL. Lactobacillus inhibitor production against Escherichia coli and coaggregation ability with uropathogens. Can. J. Microbiol. 1988;34:344–351.
- Freter M. Factors affecting the microecology of the gut. In: Fuller R, editor. Probiotics the scientific basis. Glasgow: Chapman and Hall; 1992. p. 111–145.
- Pérez PF, Minnaard Y, Disalvo EA, de Antoni GL. Surface properties of bifidobacterial strains of human origin. Appl. Environ. Microbiol. 1998;64:21–26.
- Rojas M, Conway PL. Colonization by lactobacilli of piglet small intestinal mucus. J. Appl. Bacteriol. 1996;81:474–480.
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006;2:2006–2008.
- Pilipcineca E, Huismanb TT, Willemsenb PTJ, Appelmelkc BJ, Graaf FK, Oudega B. Identification by Tn10 transposon mutagenesis of host factors involved in the biosynthesis of K99 fimbriae of Escherichia coli: Effect of LPS core mutations. FEMS Microbiol. Lett. 1994;123:201–206.
- Low D, Braaten B, van der Woude M. Fimbriae. In: Editor Neidhardt FC, Curtiss R, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE editor. Escherichia coli and Salmonella: Cellular and Molecular Biology, 2nd ed. Washington D.C.: ASM Press; 1996. Part I, Chapter 11.
- Van Molle I, Joensuu JJ, Buts L, Panjikar S, Kotiaho M, Bouckaert J, Wyns L, Niklander-Teeri V V, and De Greve H. Chloroplasts assemble the major subunit FaeG of Escherichia coli F4 (K88) fimbiae to strand-swapped dimers. J. Mol. Biol. 2007;368:791–799.
- Katakura Y, Sano R, Hashimoto T, Ninomiya K, Shioya S. Lactic acid bacteria display on the cell surface cytosolic proteins that recognize yeast mannan. Appl. Microbiol. Biotechnol. 2010;86:319–326.
