Abstract
RNA microarray analyses revealed that nuclear actin activated many human transcription factor genes including OCT4, which is required for gene reprogramming. Oct4 is known to be activated by nuclear actin in Xenopus oocytes. Our findings imply that this process of OCT4 activation is conserved in vertebrates and among cell types and could be used for gene reprogramming of human cells.
Actin is one of the most evolutionarily conserved proteins in eukaryotes. Actin exists in two functional forms, monomeric globular form (G-actin) and filamentous form (F-actin), and transition between the G-actin and F-actin is regulated by various actin-binding proteins.Citation1) In the cytoplasm, F-actin fibers are bundled up with a group of actin-binding proteins and form cytoskeleton. The formation of cytoskeleton governs various cellular functions including cell mobility and cell division.Citation2) Although actin was detected in the nucleus a long time ago, its nuclear existence was convincingly demonstrated only recently.Citation3) However, the function of nuclear actin is, so far, not fully understood.
The amount of nuclear actin is relatively low in somatic cells. However, the nucleus of oocytes contains large amount of actin. Gurdon Citation4) previously showed that somatic cell nuclei transplanted into Xenopus oocytes were subjected to gene reprogramming. Interestingly, F-actin was formed in the nuclei of mouse cells transplanted into Xenopus oocytes, and it was shown that this nuclear F-actin was required for gene reprogramming through the activation of Oct4, which is a central transcription factor for gene reprogramming.Citation5)
We recently expressed the EYFP-tagged actin (EYFP-actin) fused to a nuclear localization signal (NLS) to determine the function of the nuclear actin in cultured human cells.Citation6) Since the expression of EYFP-actin had no detectable adverse effect on actin function, this fusion protein has been widely used to monitor the subcellular distribution and function of actin in cells.Citation7) We showed that the expression of EYFP-NLS-actin led to the formation of F-actin bundles in the nucleus of a small population (1–5%) of cells, suggesting that nuclear F-actin formation is feasible.Citation6) In the course of analyzing the nuclear F-actin, we clearly observed the enhancement of transcription. However, we did not examine transcriptions of which genes were enhanced. Here, we have applied microarray analysis to elucidate the function of nuclear actin in transcription regulation.
The subcellular localizations of actin and NLS-actin in HeLa cells were determined by monitoring the fluorescence of EYFP, and the presence of nuclear F-actin was detected using the F-actin probe Lifeact,Citation8) which binds to actin oligomers and helps in visualizing F-actin both in the nucleus and cytoplasm more effectively than visualizing the F-actin bundles. Thus, the accumulation of Lifeact in the nucleus was observed in most cells expressing NLS-actin regardless of whether or not F-actin bundles were visible, but this accumulation of Lifeact in the nucleus was hardly observed in cells expressing actin (Fig. ). This result obtained using Lifeact suggests that the actin imported into the nucleus effectively formed F-actin, which is in contrast to the previous observation that the F-actin bundles derived from the nuclear actin were visible only in a small population of cells.Citation6)
Fig. 1. Expression of EYFP-NLS-actin induces the formation of nuclear F-actin.
Notes: (A) To construct the pLifeact-mCherry expression plasmid, a DNA-sequence coding for the Lifeact peptide [Citation8] was inserted into the pmCherry-N1 vector (Clonetech). The EYFP-actin and EYFP-NLS-actin expression plasmids were constructed as mentioned previously [Citation6]. HeLa cells were transiently transfected with pLifeact-mCherry together with EYFP only (top panels), EYFP-actin (middle panels), or EYFP-NLS-actin (bottom panels) expression plasmid using Fugene HD (Promega). Forty-eight hours after the transfection, HeLa cells were fixed with 4% paraformaldehyde in PBS, and cells were then permeabilized with 0.5% Triton X-100 in PBS. Cellular DNA was stained with DAPI (blue). Images were captured on a FV1000 confocal microscope system (Olympus). (B) Magnified views of images of cells shown in A. Bar = 10 μm.
![Fig. 1. Expression of EYFP-NLS-actin induces the formation of nuclear F-actin.Notes: (A) To construct the pLifeact-mCherry expression plasmid, a DNA-sequence coding for the Lifeact peptide [Citation8] was inserted into the pmCherry-N1 vector (Clonetech). The EYFP-actin and EYFP-NLS-actin expression plasmids were constructed as mentioned previously [Citation6]. HeLa cells were transiently transfected with pLifeact-mCherry together with EYFP only (top panels), EYFP-actin (middle panels), or EYFP-NLS-actin (bottom panels) expression plasmid using Fugene HD (Promega). Forty-eight hours after the transfection, HeLa cells were fixed with 4% paraformaldehyde in PBS, and cells were then permeabilized with 0.5% Triton X-100 in PBS. Cellular DNA was stained with DAPI (blue). Images were captured on a FV1000 confocal microscope system (Olympus). (B) Magnified views of images of cells shown in A. Bar = 10 μm.](/cms/asset/3b7bca11-0336-469a-8705-517d40551ce6/tbbb_a_972332_f0001_oc.gif)
We performed microarray analysis using RNAs isolated from cells expressing the EYFP-actin and from cells expressing the EYFP-NLS-actin. We then subtracted the microarray data of the former from those of the latter to eliminate any effect of cytoplasmic actin. The microarray expression profiling data were deposited in Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo) with the accession number GSE59799. Nuclear accumulation of actin resulted in up- and down-regulation of 1,293 and 811 genes (|log2 ratio| ≥ 1), respectively. The number of up-regulated genes was significantly greater than the number of down-regulated genes (p < 0.001), which is consistent with our previous observation that the nuclear F-actin enhanced transcription.Citation6)
We next focused our attention to determine the function of NLS-actin in the nucleus. For this purpose, we analyzed 11,033 gene loci containing the GO term “nucleus” (GO:0005634). The GO terms that frequently appeared (>8%) in differentially expressed up- or down-regulated genes are shown in Fig. . For two GO terms, “regulation of transcription from RNA polymerase II promoter” (GO:0006357) and “negative regulation of transcription from RNA polymerase II promoter” (GO:0000122), the number of up-regulated genes was significantly higher than the number of down-regulated genes. In contrast, for each one of the remaining GO terms, no significant difference was observed between the number of up-regulated and down-regulated genes. It is likely that these transcription factors induced by nuclear actin up-regulate and down-regulate a certain number of genes.
Fig. 2. GO terms frequently associated with differentially expressed up- and down-regulated genes as a result of expression of NLS-actin.
Notes: The microarray data was obtained from a single hybridization to Agilent SurePrint G3 Human GE v2 8 × 60 K Microarray according to the one-color labeling protocol (Agilent Technologies). Only the GO terms that appeared in more than 8% of the up- or down-regulated genes are listed. The relative appearances of GO terms for the up- and down-regulated genes are shown using filled (up-regulated genes) and open (down-regulated genes) bars, respectively. For two GO terms (GO:0006357 and GO:0000122), plots are marked with p-values, because in both cases the number of up-regulated genes associated with these two GO terms were significantly higher than the number of down-regulated genes. For the remaining GO terms, no significant difference was however observed between the number of up-regulated and down-regulated genes.
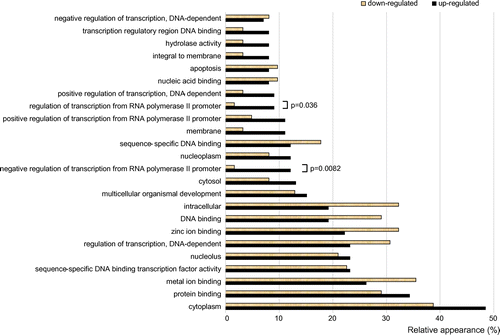
The GO terms “regulation of transcription from RNA polymerase II promoter” (GO:0006357) and “negative regulation of transcription from RNA polymerase II promoter” (GO:0000122) were associated with the following differentially expressed genes: BRD7, CDX4, CNOT2, FOXA1, FOXP3, HDAC10, HOXA2, ID4, KLF12, KLF17, MDFI, NRIP2, POU5F1 (OCT4), PSMD10, SP100, TCEA2, TCF7, CF7L1, UBASH3B, and ZNF157. Although we have future plans to elucidate the mechanism by which nuclear actin regulates each one of these transcription factor genes, in the present study, we have focused our attention only to the POU5F1 (OCT4) gene, because OCT4 is a central factor for gene reprogramming, and as described above, it has been shown to be activated by F-actin in Xenopus oocyte.Citation5) In the study of Xenopus, it was suggested that polymerized actin directly binds to Oct4 gene regions and that the binding of nuclear F-actin results in an increased chance for a chromatin remodeling complex to associate with chromatin.Citation5) We speculate about a possibility that such direct regulation of OCT4 gene by nuclear F-actin is conserved also in human cells.
Whereas the nuclear actin forms F-actin efficiently, nuclear G-actin forms heterodimer with the actin-related protein Arp4,Citation9,10) and this heterodimer is an essential component of the multiple chromatin remodeling and histone modification complexes.Citation11) Therefore, as a part of these complexes, the nuclear actin might have influence on the expression of OCT4 and other transcription factor genes listed above. However, none of these transcription factor genes, including OCT4, were found among the genes that are reported to be regulated by Apr4.Citation10) Taken together, it is likely that the expression of NLS-actin activates OCT4 and several other transcription factor genes as a result of formation of nuclear F-actin rather than the formation of actin-Arp4 heterodimer, which is part of the chromatin remodeling and histone modification complexes.
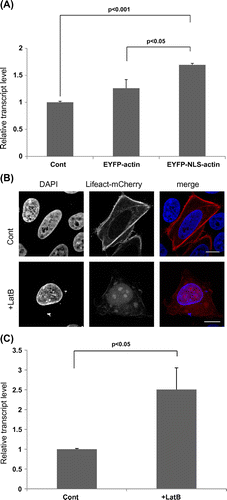
Quantitative reverse transcriptase (RT)–PCR analysis confirmed that the activation of OCT4 by the expression of NLS-actin was indeed significant. To further test the idea that the OCT4 is indeed activated by nuclear actin, we next tried increasing the level of nuclear F-actin without expressing the actin gene ectopically. It was previously shown that latrunculin B (LatB) preferentially depolymerizes cytoplasmic F-actin and increases the level of nuclear F-actin, probably by increasing the nuclear import of the cytoplasmic G-actin pool.Citation12) Accordingly, HeLa cells treated with LatB exhibited an increase in the level of nuclear F-actin, as judged by the Lifeact probe (Fig. (B)). OCT4 was also found to be significantly activated in these cells (Fig. (C)), which again supported the idea that the nuclear F-actin activates OCT4.
Fig. 3. Quantitative RT–PCR analysis of the activation of OCT4 gene induced by nuclear actin.
Notes: (A) HeLa cells transfected with EYFP-actin or EYFP-NLS-actin expression plasmid were cultured for 24 h. Total RNAs were prepared from these cells using the Qiagen RNeasy kit (Qiagen). RNAs were reverse-transcribed into cDNAs using the SuperScript III First-Strand Synthesis System (Invitrogen), and then a real-time PCR analysis for OCT4 was carried out using these cDNAs and SYBR-Green (Applied Biosystems). Values were normalized with respect to the endogenous control glyceraldehyde-3-phosphate dehydrogenase and subsequently compared to control samples using the ΔΔCt method. (B) HeLa cells expressing Lifeact-mCherry were treated with 1 μM LatB, and the presence of nuclear F-actin in these cells was observed under a microscope (lower panels). Control cells were not treated with LatB (Cont, upper panels). (C) The expression of OCT4 in control and LatB-treated cells was analyzed as in A. Data shown are mean ± SE from three independent experiments. p value was determined by student’s t-test.
In the present study, we have shown that the nuclear actin is involved in activating OCT4 and other transcription factor genes in human cultured cells. These results, taken together with the observation that the nuclear F-actin activated Oct4 in Xenopus oocytes,Citation5) suggested that this activation process is conserved in vertebrates and among cell types. OCT4 is an essential factor in creating induced pluripotent stem (iPS) cells. In fact, it has been reported that OCT4 alone is sufficient to reprogram human cells to pluripotency.Citation13) Therefore, increasing the nuclear F-actin level by expressing NLS-actin or using LatB might be a useful method for gene reprogramming and produce iPS cells. We have further future plans for exploring the role of nuclear actin in transcription and other associated practical issues.
Acknowledgments
We thank Dr Primal De Lanerolle for DNA constructs.
Additional information
Funding
Notes
Abbreviations: EYFP, enhanced yellow fluorescent protein; GO, gene ontology; iPS, induced pluripotent stem; LatB, latrunculin B.
References
- dos Remedios CG, Chhabra D, Kekic M, Dedova IV, Tsubakihara M, Berry DA, Nosworthy NJ. Actin binding proteins: regulation of cytoskeletal microfilaments. Physiol. Rev. 2003;83:433–473.
- Pollard TD, Cooper JA. Actin, a central player in cell shape and movement. Science. 2009;326:1208–1212.10.1126/science.1175862
- Simon DN, Wilson KL. The nucleoskeleton as a genome-associated dynamic ‘network of networks’. Nat. Rev. Mol. Cell. Biol. 2011;12:695–708.10.1038/nrm3207
- Gurdon JB. The developmental capacity of nuclei taken from intestinal epithelium cells of feeding tadpoles. J. Embryol. Exp. Morphol. 1962;10:622–640.
- Miyamoto K, Pasque V, Jullien J, Gurdon JB. Nuclear actin polymerization is required for transcriptional reprogramming of Oct4 by oocytes. Genes Dev. 2011;25:946–958.10.1101/gad.615211
- Kalendová A, Kalasová I, Yamazaki S, Uličná L, Harata M, Hozák P. Nuclear actin filaments recruit cofilin and actin-related protein 3, and their formation is connected with a mitotic block. Histochem. Cell Biol. 2014;142:139–152.10.1007/s00418-014-1243-9
- Herget-Rosenthal S, Hosford M, Kribben A, Atkinson SJ, Sandoval RM, Molitoris BA. Characteristics of EYFP-actin and visualization of actin dynamics during ATP depletion and repletion. Am. J. Physiol. Cell Physiol. 2001;281:C1858–1870.
- Riedl J, Crevenna AH, Kessenbrock K, Yu JH, Neukirchen D, Bista M, Bradke F, Jenne D, Holak TA, Werb Z, Sixt M, Wedlich-Soldner R. Lifeact: a versatile marker to visualize F-actin. Nat. Methods. 2008;5:605–607.10.1038/nmeth.1220
- Fenn S, Gerhold CB, Hopfner KP. Nuclear actin-related proteins take shape. Bioarchitecture. 2011;1:192–195.
- Nishimoto N, Watanabe M, Watanabe S, Sugimoto N, Yugawa T, Ikura T, Koiwai O, Kiyono T, Fujita M. Heterocomplex formation by Arp4 and β-actin is involved in the integrity of the Brg1 chromatin remodeling complex. J. Cell Sci. 2012;125:3870–3882.10.1242/jcs.104349
- Oma Y, Harata M. Actin-related proteins localized in the nucleus: from discovery to novel roles in nuclear organization. Nucleus. 2011;2:38–46.10.4161/nucl
- Belin BJ, Cimini BA, Blackburn EH, Mullins RD. Visualization of actin filaments and monomers in somatic cell nuclei. Mol. Biol. Cell. 2013;24:982–994.10.1091/mbc.E12-09-0685
- Kim JB, Greber B, Araúzo-Bravo MJ, Meyer J, Park KI, Zaehres H, Schöler HR. Direct reprogramming of human neural stem cells by OCT4. Nature. 2009;461:649–653.10.1038/nature08436
