Abstract
Diadenosine 5′,5′′′-P1,P4-tetraphosphate (Ap4A) phosphorylase from Mycobacterium tuberculosis H37Rv (MtAPA) belongs to the histidine triad motif (HIT) superfamily, but is the only member with an alanine residue at position 149 (Ala-149). Enzymatic analysis revealed that the Ala-149 deletion mutant displayed substrate specificity for diadenosine 5′,5′′′-P1,P5-pentaphosphate and was inactive on Ap4A and other substrates that are utilized by the wild-type enzyme.
Key words:
Mycobacterium tuberculosis is the causative agent of tuberculosis (TB). In 2012, approximately 1.3 million people died from TB worldwide, and more than 8.6 million people developed TB. Furthermore, multidrug-resistant and extensively drug-resistant TB strains pose a growing public health threat and economic burden globally (World Health Organization, http://www.who.int/tb/publications/global_report/en/). This has created an urgent need to discover novel anti-TB drugs. To facilitate the structure-based design of new anti-TB drugs, our group has focused on a novel diadenosine 5′,5′′′-P1,P4-tetraphosphate (Ap4A) phosphorylase (EC 2.7.7.53), which converts Ap4A to ATP and ADP in the presence of inorganic phosphate, from M. tuberculosis H37Rv (MtAPA).Citation1,2) Rv2613c gene, which encodes MtAPA, is an essential gene in M. tuberculosis H37Rv,Citation3) and in silico analyses have shown that MtAPA could be a target for new anti-TB drugs.Citation4) Mutations in an Ap4A degradation enzyme lead to Ap4A accumulation in cells, which in turn causes defects in transcription, sigmaF-mediated gene regulation, and catabolite repression.Citation5) Therefore, the development of specific MtAPA inhibitors could lead to production of a novel series of anti-TB drugs.
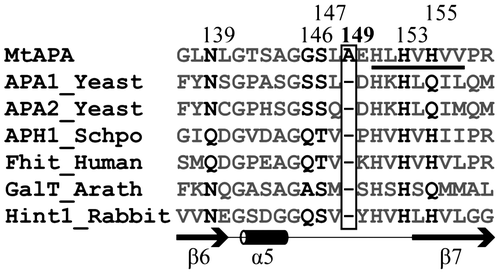
To facilitate the design of specific MtAPA inhibitors, we previously determined the three-dimensional structure of MtAPA and elucidated structure–function relationships.Citation2) Since the amino acid sequence of MtAPA has a histidine triad (HIT) motif (His-φ-His-φ-His-φ-φ, where φ represents a hydrophobic amino acid) (Fig. ), the enzyme belongs to the HIT superfamily. This superfamily also includes nucleotide hydrolases and transferases, for example, Saccharomyces cerevisiae Ap4A phosphorylases 1 and 2 (APA1_Yeast and APA2_Yeast, respectively) and Homo sapiens HIT family ApnA hydrolase (Fhit_Human) (Fig. and Citation6)). In addition to MtAPA, the crystal structures of some HIT superfamily proteins have been reported.Citation7–11) The overall and active site structures of MtAPA are similar to those of other HIT superfamily proteins.Citation2) However, some amino acid residues that contribute to the active site of MtAPA are divergent from those of other HIT superfamily proteins (Fig. ). In particular, an Ala residue at position 149 (Ala-149) is unique to MtAPA in comparison with all other HIT superfamily members (Fig. ). Furthermore, Ala-149 is situated within a flexible loop that is located at the active site of MtAPA, suggesting that it could play a crucial role in modulating catalytic activity and substrate specificity of this enzyme.
Fig. 1. Multiple sequence alignment of the active site region of MtAPA and HIT superfamily proteins (S. cerevisiae Ap4A phosphorylases 1 and 2; APA1_Yeast and APA2_Yeast, Schizosaccharomyces pombe HIT family ApnA hydrolase; APH1_Schpo, H. sapiens HIT family ApnA hydrolase; Fhit_Human, Arabidopsis thaliana galactose-1-phosphate uridylyltransferases; GalT_Arath, and Oryctolagus cuniculus AMP-lysine hydrolase; Hint1_Rabbit).
Notes: Sequence alignment was performed using the ClustalW 2.1 software.Citation12) The HIT motif is underlined. The numbers along the top refer to the positions of amino acids in the sequence of MtAPA. The Ala-149 in MtAPA and the corresponding positions in other HIT superfamily proteins are shown in the box. In addition to Ala-149, the amino acids corresponding to Asn-139, Gly-146, Ser-147, His-153, and His-155 in MtAPA are shown in black, since these amino acids were previously presented as important residues for enzymatic activity of MtAPA,Citation2) and other amino acids are gray. The secondary structure of the active site based on the crystal structure of MtAPA is shown. The SWISS-PROT or TrEMBL accession numbers are as follows: MtAPA (P9WMK9), APA1_Yeast (P16550), APA2_Yeast (P22108), APH1_SCHPO (P49776), Fhit_Human (P49789), Galt_Arath (Q9FK51), and Hint1_Rabbit (P80912).
The Ala-149 deletion was introduced into Rv2613c using the QuickChange Site-Directed Mutagenesis Kit II (Agilent Technologies, Inc., Santa Clara, CA) and the plasmid pMS2613c, which was previously constructed,Citation1) as the template. The Ala-149 deletion primer (5′-ggcgggtcgctggagcacctgcac-3′) and the Ala-149 deletion antisense primer (5′-gtgcaggtgctccgcgacccgcc-3′) were designed using QuickChange Primer Design Program (Agilent Technologies). The resulting plasmid was sequenced with an Applied Biosystems 3130xl Genetic Analyzer (Applied Biosystems, Carlsbad, CA) to confirm the mutations; verified DNAs were used to transform Escherichia coli BL21(DE3)pLysS (Promega, Woods Hollow Road Madison, WI) for overexpression of the Ala-149 deletion construct (Δ149A-MtAPA). The Δ149A-MtAPA was purified to homogeneity by a two-step column chromatography, as described previously.Citation1) Analysis of gel filtration chromatography indicated that the molecular weight of the Δ149A-MtAPA is approximately 100 kDa (data not shown), indicating that this substituent formed a homotetramer of 25-kDa subunits in solution, which was also observed with wild-type MtAPA. Because the tetrameric structure of wild-type MtAPA is reported to be essential for the formation of the Ap4A-binding site,Citation2) we believe that the substrate-binding site of Δ149A-MtAPA is also closely linked to the tetrameric structure.
The enzyme activity of wild-type MtAPA and Δ149A-MtAPA was measured using a high-performance liquid chromatography system (Shimadzu, Kyoto, Japan) to quantify the amount of substrate remaining after the reaction as described previously.Citation1) The kinetic parameters of wild-type MtAPA and Δ149A-MtAPA were then compared. The Km and kcat values for Ap4A of Δ149A-MtAPA were 0.19 ± 0.048 mM and 1.25 ± 0.099 s−1, respectively. On the other hand, the Km and kcat values for Ap4A of wild-type MtAPA were 0.10 ± 0.001 mM and 8.48 ± 0.313 s−1, respectively (Supplemental Fig. 1).Citation1) Furthermore, the kcat/Km value for Ap4A of Δ149A-MtAPA (6.58 mM−1 s−1) was approximately 10% of that of wild-type MtAPA (84.8 mM−1 s−1).
The substrate specificities of wild-type MtAPA and Δ149A-MtAPA were then compared. Although wild-type MtAPA could use a variety of substrates, Δ149A-MtAPA showed diadenosine 5′,5′′′-P1,P5-pentaphosphate (Ap5A) specificity (Table ). When wild-type MtAPA activity was further examined using various substrates, Ap5A, Ap4G, Gp4G, and Gp5G were phosphorylated with approximately the same efficiency as Ap4A (Table ). Furthermore, Ap5G, Ap4U, Ap5U, and Ap5dT were phosphorylated with approximately half the efficiency of Ap4A (Table ). On the other hand, the phosphorylation of all the dinucleotide polyphosphates by the Δ149A-MtAPA mutant was less than 50% compared to that of the Ap5A substrate (Table ). Moreover, the Km and kcat values for Ap5A of Δ149A-MtAPA were 0.03 ± 0.006 mM and 2.21 ± 0.092 s−1, respectively. On the other hand, the Km and kcat values for Ap5A of wild-type MtAPA were 0.07 ± 0.007 mM and 8.29 ± 0.218 s−1, respectively (Supplemental Fig. 1). Therefore, the kcat/Km value yielded by Δ149A-MtAPA with Ap5A as a substrate (73.7 mM−1 s−1) was approximately 10-fold higher than that for Ap4A (6.58 mM−1 s−1), but this difference was not observed when wild-type MtAPA was tested (118.4 mM−1 s−1 for Ap5A and 84.8 mM−1 s−1 for Ap4A).
Table 1. Nucleotide substrate utilization by wild-type MtAPA and ∆149A-MtAPA.
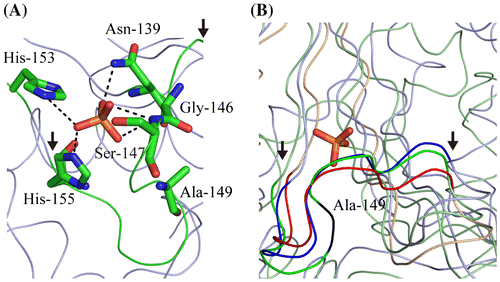
Ala-149 is present on a loop that is located in the active site of MtAPA and does not appear to directly impact catalytic activity (Fig. (A)). The deletion of Ala-149 leads to shortening of the loop. Because other HIT superfamily proteins do not have the corresponding Ala-149 residue (Fig. ), their loops are likely shorter, as exemplified by other HIT superfamily proteins (Fig. (B)). Furthermore, other HIT superfamily proteins do not show broad substrate profiles such as MtAPA; for example, the activities of APA1_Yeast and APA2_Yeast with Ap4A are higher than those observed with Ap3A and Ap5A,Citation7) and Fhit_Human has higher activity toward Ap3A when compared to other dinucleotide polyphosphates.Citation13) This has led to the suggestion that the flexibility of this loop might be correlated with substrate specificity. Therefore, the flexibility of this loop could be useful for designing MtAPA-specific inhibitors, which bind the substrate-binding site of MtAPA.
Fig. 2. Ribbon model of the active site of MtAPA (A) and the comparison of the loop conformation within it (B).
Notes: Ribbon diagrams were generated by PyMOL (W.L. DeLano, http://www.pymol.org). The atomic coordinates of crystal structures were downloaded from PDB (www.rcsb.org). It is likely that the phosphate ion-binding site of MtAPA (PDB ID; 3ANO) is the bona fide active site. In this site, Asn-139, Ser-147, His-153, and His-155 coordinate the phosphate ion, whereas Gly-146 is in close proximity.Citation2) (A) The loop in the active site is shown in green, with the start and end of the loop indicated by arrows. Asn-139, Gly-146, Ser-147, Ala-149, His-153, His-155, and the phosphate ion are represented by sticks. Carbon, nitrogen, oxygen, and phosphorus atoms are shown in green, blue, red, and orange, respectively. (B) The ribbon models of the active site of MtAPA, Fhit_Human (PDB ID; 6FIT), and GalT_Arath (PDB ID; 1Z84) are shown in green, blue, and red, respectively. The position of Ala-149 in MtAPA is shown in black. The phosphate ion in the active site of MtAPA is represented by sticks. The positions of the start and end of the loop in the active site are indicated by arrows.
Conflicts of interest
There are no conflicts of interest to declare.
Supplemental material
The supplemental material for this paper is available at http://dx.doi.org/10.1080/09168451.2014.973364.
Supplemental Materials
Download MS Power Point (208.5 KB)Additional information
Funding
References
- Mori S, Shibayama K, Wachino J, Arakawa Y. Purification and molecular characterization of a novel diadenosine 5′,5′′′-P1,P4-tetraphosphate phosphorylase from Mycobacterium tuberculosis H37Rv. Protein Express. Purif. 2010;69:99–105.10.1016/j.pep.2009.09.010
- Mori S, Shibayama K, Wachino J, Arakawa Y. Structural insights into the novel diadenosine 5′,5‴-p1,p4-tetraphosphate phosphorylase from Mycobacterium tuberculosis H37Rv. J. Mol. Biol. 2011;410:93–104.10.1016/j.jmb.2011.04.059
- Sassetti CM, Boyd DH, Rubin EJ. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 2003;48:77–84.10.1046/j.1365-2958.2003.03425.x
- Raman K, Yeturu K, Chandra N. targetTB: a target identification pipeline for Mycobacterium tuberculosis through an interactome, reactome and genome-scale structural analysis. BMC Syst. Biol. 2008;2:109.10.1186/1752-0509-2-109
- Farr SB, Arnosti DN, Chamberlin MJ, Ames BN. An apaH mutation causes AppppA to accumulate and affects motility and catabolite repression in Escherichia coli. Proc. Nat. Acad. Sci. USA. 1989;86:5010–5014.10.1073/pnas.86.13.5010
- Brenner C. Hint, Fhit, and GalT: function, structure, evolution, and mechanism of three branches of the histidine triad superfamily of nucleotide hydrolases and transferases. Biochemistry. 2002;41:9003–9014.10.1021/bi025942q
- Hou WT, Li WZ, Chen Y, Jiang YL, Zhou CZ. Structures of yeast Apa2 reveal catalytic insights into a canonical Ap4A phosphorylase of the histidine triad superfamily. J. Mol. Biol. 2013;425:2687–2698.10.1016/j.jmb.2013.04.018
- Lima CD, Klein MG, Hendrickson WA. Structure-based analysis of catalysis and substrate definition in the hit protein family. Science. 1997;278:286–290.10.1126/science.278.5336.286
- Pace HC, Garrison PN, Robinson AK, Barnes LD, Draganescu A, Rosler A, Blackburn GM, Siprashvili Z, Croce CM, Huebner K, Brenner C. Genetic, biochemical, and crystallographic characterization of Fhit–substrate complexes as the active signaling form of Fhit. Proc. Nat. Acad. Sci. USA. 1998;95:5484–5489.10.1073/pnas.95.10.5484
- Krakowiak A, Pace HC, Blackburn GM, Adams M, Mekhalfia A, Kaczmarek R, Baraniak J, Stec WJ, Brenner C. Biochemical, crystallographic, and mutagenic characterization of hint, the amp-lysine hydrolase, with novel substrates and inhibitors. J. Biol. Chem. 2004;279:18711–18716.10.1074/jbc.M314271200
- McCoy JG, Arabshahi A, Bitto E, Bingman CA, Ruzicka FJ, Frey PA, Phillips GN Jr. Structure and mechanism of an adp-glucose phosphorylase from Arabidopsis thaliana. Biochemistry. 2006;45:3154–3162.10.1021/bi052232m
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948.10.1093/bioinformatics/btm404
- Barnes LD, Garrison PN, Siprashvili Z, Guranowski A, Robinson AK, Ingram SW, Croce CM, Ohta M, Huebner K. Fhit, a putative tumor suppressor in humans, is a dinucleoside 5′,5′′′-P1, P3-triphosphate hydrolase. Biochemistry. 1996;35:11529–11535.10.1021/bi961415t
