Abstract
A protein kinase C of Aspergillus nidulans, PkcA, is required for cell wall integrity (CWI) and is considered a major component of the regulating pathway. To investigate whether PkcA regulates the transcription of cell wall-related genes, we constructed strains expressing pkcA(R429A) that encodes an activated form of PkcA. The mRNA levels of most chitin synthase genes and an α-glucan synthase gene, agsB, were increased when pkcA(R429A) expression was induced. These mRNA increases were not observed or were only partially observed, in a deletion mutant of rlmA, an ortholog of RLM1 that encodes a transcription factor in the CWI pathway in Saccharomyces cerevisiae. In addition, in a pkcA temperature-sensitive mutant under heat stress, the mRNA levels of some chitin synthase genes and agsB did not increase. These results suggest that PkcA is involved in CWI maintenance through the transcriptional regulation of cell wall-related genes.
Graphical Abstract
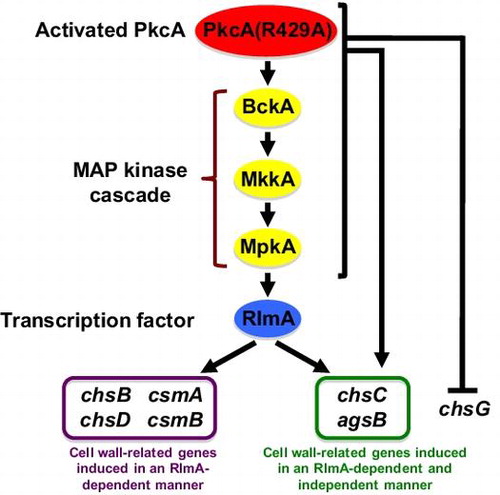
Signal transduction pathway that regulates the transcription of cell wall-related genes by an activated PKC in Aspergillus nidulans
The fungal cell wall is a highly elaborate, rigid structure that is essential for morphologenesis and protection against adverse environmental conditions. It is well known that the cell wall is remodeled in response to environmental changes of external environmental conditions. It consists mainly of polysaccharides, α-glucan, β-glucan, and chitin in filamentous fungi.Citation1) Aspergillus nidulans has two α-1,3-glucan synthase genes, agsA and agsB, a β-1,3-glucan synthase gene, fksA, and eight chitin synthase genes, chsA, chsB, chsC, chsD, chsF, chsG, csmA, and csmB.Citation2,3) AgsB functions as the main α-1,3-glucan synthase and has been implicated in the cell wall stress response.Citation4,5) The β-1,3-glucan synthase gene in Aspergillus fumigatus has been shown to be essential for growthCitation6); therefore, it is likely that fksA is also essential for the growth of A. nidulans. chsB plays a crucial role in hyphal growth, and it has been implicated in septum formation.Citation7–9) chsD is important for hyphal growth and morphology when chsB expression is repressed.Citation10) It has been suggested that CsmA and CsmB function in hyphal growth and that their importance increases under low osmotic conditions.Citation11,12) chsA and chsC show significant functional redundancy and are thought to be directly involved in septum formation.Citation13,14) The functions of chsF and chsG are currently unknown.
Protein kinase C (PKC) is a serine/threonine kinase that is conserved from yeasts to mammals and plays pivotal roles in various signal transduction processes. The budding yeast Saccharomyces cerevisiae has only one PKC-encoding gene, PKC1, and its gene product, Pkc1p, is thought to be involved in various cellular processes.Citation15,16) A PKC1 deletion mutant did not grow under normal conditions; however, it did grow on medium with osmotic stabilizers.Citation15) One of the main functions of Pkc1p is signal transduction in the cell wall integrity (CWI) pathway. When cells are exposed to cell wall stress, Pkc1p is activated and it phosphorylates Bck1p, which activates a mitogen-activated protein (MAP) kinase cascade. This MAP kinase cascade includes Bck1p, Mkk1p and Mkk2p, and Slt2p, as a MAP kinase kinase kinase, two MAP kinase kinases, and a MAP kinase, respectively. Slt2p activates a transcription factor, Rlm1p, and a transcription regulatory complex, the SBF complex, which is composed of Swi4p and Swi6p. Eighteen genes were shown to be induced by the activation of the CWI pathway, and most of these genes encoded either integral cell wall proteins or enzymes involved in cell wall biosynthesis.Citation17) In addition, the consensus sequence recognized by Rlm1p (TA(T/A)4TAG) or similar sequences were found in the promoter regions of all 18 genes,Citation17) and the SBF complex has been shown to regulate the expression of the β-1,3-glucan synthase genes FKS1 and FKS2.Citation16)
In A. nidulans, a single PKC-encoding gene, pkcA, was isolated and characterized. pkcA is essential for viability even when grown on medium supplemented with osmotic stabilizers.Citation18) Repression of pkcA expression leads to hypersensitivity to cell wall-perturbing agents and cell wall structure defects.Citation19,20) In addition, A. nidulans has orthologs of most CWI pathway components.Citation4) These findings suggest that the CWI pathway in yeasts is also present in A. nidulans and that PkcA is involved in this pathway. PkcA was shown to be localized to the hyphal apices, forming septa, and tips of phialides.Citation20) PkcA is thought to be involved in conidiation, germination, penicillin production, unfolded-protein response, and farnesol-induced cell death.Citation18–23) Previously, we constructed a temperature-sensitive pkcA mutant (pkcA-ts) and showed that this mutant does not grow at 42 °C due to PkcA inactivation.Citation24) Based on our findings using this mutant, we suggested that PkcA is involved in suppression of apoptosis and polarity establishment under heat stress.Citation24)
In A. nidulans, the transcription of agsA and agsB was shown to be regulated by MpkA and RlmA, which are orthologs of Slt2p and Rlm1p, respectively; however, transcription of fksA and the chitin synthase genes was not dependent on MpkA and RlmA when the cells were treated with the β-1,3-glucan synthase inhibitor micafungin.Citation4) In contrast, a recent report showed that treatment with micafungin induced fksA and csmB expression in the wild-type strain, but not in an mpkA deletion mutant. In addition, the mRNA level of chsB in the mpkA deletion mutant decreased under micafungin treatment.Citation25) Therefore, whether MpkA and RlmA regulate the transcription of fksA and the chitin synthase genes is controversial.
In this study, to determine whether PkcA is involved in the transcriptional regulation of cell wall-related genes, we constructed strains that produce an activated form of PkcA. The results of our transcriptional analyses of cell wall-related genes suggest that PkcA is involved in the transcriptional regulation of most the chitin synthase genes and agsB.
Materials and methods
Strains and growth conditions
The A. nidulans strains used in this study are listed in Table . Minimal medium (MMG) was prepared as described by Rowlands and Turner.Citation26) In MMG, 2% glucose was added as a sole carbon source. In MMTF, 100 mM threonine and 0.1% fructose were added as carbon sources instead of glucose in MMG. In MMT, 100 mM threonine was added as a carbon source instead of glucose in MMG. YG medium (0.5% yeast extract, 1% glucose, 0.1% trace elementsCitation26)) was also used. MMTF and MMT were used to induce the expression of alcA promoter. For the growth of pyroA4, riboB2, and pyrG89 mutants, 0.5 μg/mL pyridoxine, 2.5 μg/mL riboflavin, and 10 mM uridine and 10 mM uracil, respectively, were added to MMG, MMTF, or MMT. Solid media were prepared by adding 1.5% agar. Bacterial and fungal transformations were performed as described previously.Citation10)
Table 1. A. nidulans strains used in this study.
Plasmid constructs and oligonucleotides
The primers used for the construction of plasmids are listed in Table S1. To create the R429A mutants, we constructed plasmids pBSriboB and palcA(p)-R429A as follows: A 3.0-kb fragment containing riboB was amplified from the total DNA of the strain A1149 using a primer set of 5riboBn and 3riboBn. The amplified fragment was digested with NotI and XhoI and ligated with NotI- and XhoI-digested pBluescript II SK+ (Stratagene) to yield pBSriboB. We confirmed that no mutation was introduced into the sequence in the cloned PCR product. To construct palcA(p)-R429A, we used a double-joint PCR method.Citation27) A 4.7-kb fragment containing the upstream of pkcA, the entire gene of riboB, alcA promoter, and the coding region of the N-terminus of PkcA was amplified from the total DNA of the strain alcA(p)-pkcA-3Citation24) using a primer set of 5pkcAF and pkcAR429AR. A 3.5-kb fragment containing the coding region of the C-terminus of PkcA and the downstream of pkcA was amplified from the total DNA of alcA(p)-pkcA-3 using a primer set of 3pkcAR and pkcAR429AF. These amplified fragments were fused by the double-joint PCR using a primer set of 5npkcAF and 3pkcAR. A 6.0-kb NdeI fragment of this fused fragment was ligated with a 5.5-kb NdeI fragment of pBSriboB, yielding palcA(p)-R429A.
To create the rlmA deletion mutants, we constructed plasmids pUCpyroA2 and pΔrlmA as follows: A 2.8-kb fragment containing the entire gene of pyroA was amplified from the total DNA of the strain A26 using a primer set of pyroA-41 f.PstI and pyroA-2684r. The amplified fragment was digested with BamHI and PstI and ligated with BamHI- and PstI-digested pUC18 (TaKaRa) to yield pUCpyroA2. A 1.0-kb fragment containing an upstream of rlmA was amplified from the total DNA of A1149 using a primer set of 5rlmAF and 5pA-rlmAR. A 1.4-kb fragment containing a downstream of rlmA was amplified from the total DNA of A1149 using a primer set of 3pA-rlmAF and 3rlmAR. A 2.4-kb fragment containing pyroA was amplified from pUCpyroA2 using a primer set of pyroA5n and pyroA3n. These amplified fragments were fused by the double-joint PCR using a primer set of 5rlmAF and 3rlmAR. A 3.9-kb EcoRI fragment of this fused fragment was ligated with the EcoRI-digested pUC18, yielding pΔrlmA.
Construction of A. nidulans strains by transformation
We constructed the strains expressing pkcA(R429A) under the control of alcA promoter as follows: A1145 was transformed with a 7.8-kb SpeI-XhoI fragment of palcA(p)-R429A (Fig. S1A). The total DNA of the transformants was extracted as described previously,Citation28) and Southern blot analysis was performed as described previously.Citation29) The two transformants in which a single copy of the transformed fragment was inserted at the riboB locus confirmed by Southern blot analysis were designated R429A-1 and R429A-2 (Fig. S1B). Since they exhibited the same phenotypes under the conditions tested, we chose R429A-1 for further analysis.
We constructed the rlmA deletion mutants as follows: A1149 and R429A-1 were transformed with a 3.9-kb EcoRI fragment of pΔrlmA (Fig. S1C). Two transformants obtained from each transformation in which rlmA was replaced with a single copy of the transformed fragment at rlmA locus confirmed by Southern blot analysis were designated ΔrlmA-1 and ΔrlmA-2 (Fig. S1D), and R429AΔrlmA-1 and R429AΔrlmA-2 (Fig. S1E). Since ΔrlmA-1 and ΔrlmA-2, and R429AΔrlmA-1 and R429AΔrlmA-2 exhibited the same phenotypes under the conditions tested, we chose ΔrlmA-1 and R429AΔrlmA-1 for further analysis. The strains R429ApyroA-1 and R429ApyroA-2 were constructed by transformation of R429A-1 with a 1.4-kb BglII-MluI fragment of pUCpyroA2. Since R429ApyroA-1 and R429ApyroA-2 exhibited the same phenotypes under the conditions tested, we chose R429ApyroA-1 for further analysis.
Total RNA extraction and RT-PCR
Total RNAs were extracted from mycelia using an RNAeasy Mini Kit (Qiagen). Reverse transcription of RNA was performed using a PrimeScript™ RT reagent Kit (Perfect Real Time) (TaKaRa) according to the manufacturer’s instructions. Real-time quantitative PCR was performed with primers listed in Table S2 using SYBR Premix Ex Taq (Perfect Real Time) (TaKaRa) according to the manufacturer’s instructions.
Preparation of cell lysate and western blot analysis
Preparation of total cell extracts and Western blot analysis were performed as described previouslyCitation24) with slight modifications. TNE buffer (20 mM Tris-HCl, 2 mM EDTA, 150 mM NaCl, pH 7.4) was used as an extraction buffer. Samples were separated by electrophoresis on 10% polyacrylamide gel containing sodium dodecyl sulfateCitation30) and electroblotted to Biotrace™ Nitrocellulose transfer membrane (Pall Corporation). To detect MpkA, the membranes were incubated with a mouse anti-p44/42 MAPK (Erk1/2) (L3F12) antibody (Cell Signaling Technology) as a primary antibody at a 1:1000 dilution and with a horseradish peroxidase (HRP)-conjugated anti-mouse IgG antibody (Cell Signaling Technology) as a secondary antibody at a 1:5000 dilution. To detect phosphorylated MpkA, the membranes were incubated with a rabbit anti-phospho-p44/42 MAPK antibody (Cell Signaling Technology) as a primary antibody at a 1:1000 dilution and with HRP-conjugated anti-rabbit IgG antibody (Cell Signaling Technology) as a secondary antibody at a 1:10,000 dilution.
Determination of conidiation efficiency
Conidiation efficiency was determined as follows: 103 conidia were spotted on the MMG plate or the MMTF plate and incubated for 72 h. All conidia were harvested from the colony and counted the number. The number of conidia divided by the colony diameter was defined as the conidiation efficiency.
Results
Construction and characterization of strains producing an activated form of PkcA
To investigate the function of PkcA, we constructed strains that produce an activated form of PkcA. In S. cerevisiae, Pkc1pR398A, in which Arg398 locating within the pseudosubstrate sequence that negatively regulates PKC activity is replaced with Ala, behaves as an activated form of Pkc1p.Citation31) Since Arg398 of Pkc1p is conserved in the amino acid sequence of PkcA in A. nidulans (as Arg429), we constructed and characterized a strain, R429A-1, that ectopically produces a mutant PkcA, PkcA(R429A), in which Arg429 was replaced with Ala, under the control of the alcA promoter (see Materials and methods). The colony diameter of R429A-1 was nearly the same as that of the wild-type strain under the pkcA(R429A)-repressing condition; however, under the pkcA(R429A)-inducing condition, the colony diameter of R429A-1 was approximately 80% of that of the wild-type strain (Fig. (A)). This suggests that overproduction of PkcA(R429A) is detrimental to the growth of A. nidulans. In contrast, the conidiation efficiency of R429A-1 was less than one-half that of the wild-type strain under the pkcA(R429A)-repressing condition, while under the pkcA(R429A)-inducing condition, the conidiation efficiency of R429A-1 was nearly the same as that of the wild-type strain (Fig. (B)). The hyphae of R429A-1 frequently formed split tips and branches close to the tips (Fig. (C)). In addition, a brown pigment was observed in the medium in R429A-1 under the pkcA(R429A)-inducing condition (data not shown). To investigate the change in PkcA activity under the pkcA(R429A)-repressing and pkcA(R429A)-inducing conditions, we analyzed the phosphorylation levels of MpkA in these strains. Under the pkcA(R429A)-repressing condition, MpkA phosphorylation level in R429A-1 was similar to that in the wild-type strain (Fig. (D)). The MpkA phosphorylation level in the alcA(p)-pkcA-3 strain, which expresses wild-type pkcA under the control of the alcA promoterCitation24), was 2.01-fold higher than that in the wild-type strain under the pkcA-inducing condition, while that in R429A-1 was 3.34-fold higher under the same condition (Fig. (E)). This indicates that PkcA(R429A) acts as an activated form of PkcA in A. nidulans.
Fig. 1. Characterization of R429A-1.
Notes: (A) Growth of R429A-1. 103 conidia of the wild-type strain (A1149) and R429A-1 were inoculated on MMG and MMTF, which are pkcA(R429A)-repressing and pkcA(R429A)-inducing conditions, respectively, and incubated for 72 h at 37 °C. (B) Conidiation efficiency of R429A-1. Conidia of the wild-type strain and R429A-1 were inoculated and incubated as described above, and the number of conidia was measured. Data are shown as means ± S.E.M. of three independent experiments. (C) Hyphal tip morphologies of the wild-type strain and R429A-1 incubated on MMTF for 72 h at 37 °C. Bars, 10 μm. (D, E) Phosphorylation levels of MpkA under the pkcA(R429A)-repressing (D) or pkcA(R429A)-inducing (E) condition. Cell extracts were prepared from the wild-type strain, alcA(p)-pkcA-3, and R429A-1, which were grown on MMG (D) or MMTF (E) for 20 h at 37 °C. The upper panels indicate the phosphorylated MpkA (ⓟ-MpkA) and the lower panels indicate the MpkA (MpkA). The numbers under the lower panels indicate the ratio of MpkA phosphorylation levels.
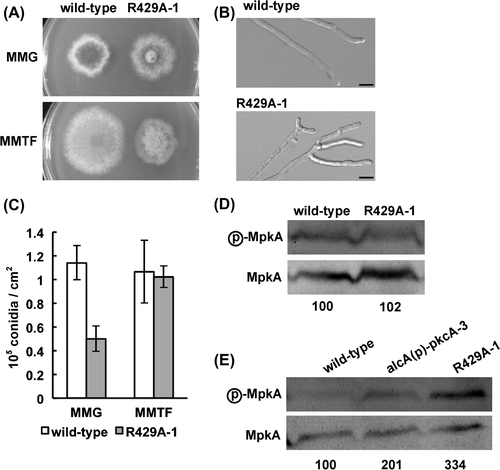
It was reported that overexpression of pkcA led to increased sensitivity to Congo red and micafungin.Citation18) In agreement with the findings in that study, alcA(p)-pkcA-3 was hypersensitive to Congo red under the pkcA-inducing condition, but was not sensitive to 1 ng/mL micafungin (Fig. ). Although under the pkcA(R429A)-inducing condition, R429A-1 was highly sensitive to calcofluor white and Congo red, it was more resistant to micafungin than the wild-type and alcA(p)-pkcA-3 strains (Fig. ).
Fig. 2. Sensitivity to cell wall-perturbing agents of R429A-1 under the pkcA(R429A)-inducing condition.
Notes: 103 conidia of the wild-type strain (A1149), alcA(p)-pkcA-3, and R429A-1 were inoculated on MMTF and MMTF containing calcofluor white (CFW), Congo red (CR), or micafungin, and incubated for 72 h at 37 °C.
Expression of cell wall-related genes under the pkcA(R429A)-inducing condition
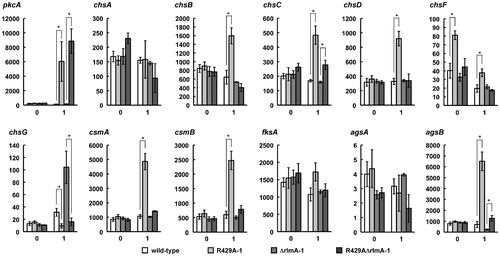
To investigate the involvement of PkcA in the transcriptional regulation of cell wall-related genes, we measured their mRNA levels by real-time quantitative PCR in R429A-1 grown both under the pkcA(R429A)-repressing condition and after shifting to the pkcA(R429A)-inducing condition. The mRNA levels of chsB, chsC, chsD, chsF, csmA, csmB, and agsB were 2.48-, 2.86-, 2.80-, 1.93-, 4.52-, 4.11-, and 9.32-fold higher, respectively, in R429A-1 than in the wild-type strain when incubated for 1 h under the pkcA(R429A)-inducing condition (Fig. ). Since the mRNA level of chsF in R429A-1 was 2.02-fold higher than that in the wild-type strain before the shift, it is unclear whether PkcA regulates the transcription of chsF (Fig. ). In contrast, although the mRNA level of chsG in the wild-type strain increased after the shift, such an increase was not detected in R429A-1 (Fig. ). These results suggest that PkcA is involved in the transcriptional upregulation of chsB, chsC, chsD, csmA, csmB, and agsB and the downregulation of chsG.
Fig. 3. mRNA levels of the cell wall-related genes 1 h after shift to the pkcA(R429A)-inducing condition.
Notes: Total RNAs were prepared from the wild-type strain (A1149), R429A-1, ΔrlmA-1, and R429AΔrlmA-1, which were grown in MMG for 20 h at 37 °C and then were incubated in MMT for 1 h. The x-axes indicate the incubation time in MMT. The y-axes indicate the copy number of mRNA per 1 pg total RNA. Data are shown as means ± S.E.M. of three independent experiments. Statistical significance of data is given by the p-value (*p < 0.05).
The role of RlmA in the expression of cell wall-related genes
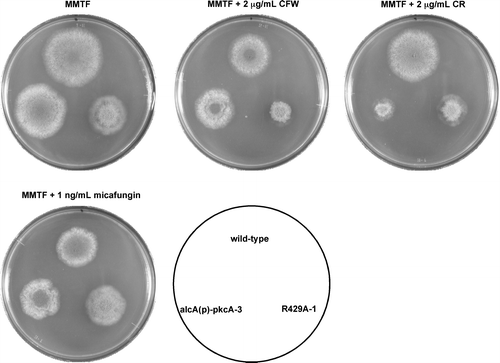
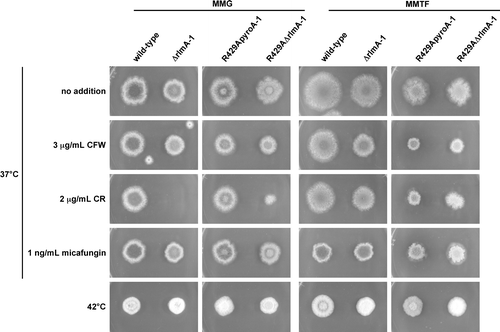
In S. cerevisiae, Rlm1p is involved in the transcriptional upregulation of cell wall-related genes, including a chitin synthase gene, CHS3, and a β-1,3-glucan synthase gene, FKS1.Citation17) It has been suggested that when treated with micafungin, the increase in the mRNA levels of fksA and the chitin synthase genes in A. nidulans is not dependent on RlmA.Citation4) However, sequences similar to the consensus Rlm1p-binding sequence are present in the promoter region between csmA and csmBCitation12) and in the promoter regions of chsB, chsC, chsD, and agsB (Table ). Therefore, it is possible that PkcA regulates the transcription of these genes through RlmA. To examine this possibility, we constructed two rlmA deletion mutant strains, ΔrlmA-1 and R429AΔrlmA-1, the latter of which expresses pkcA(R429A) under the control of the alcA promoter in addition to the wild-type pkcA. ΔrlmA-1 exhibited a slight growth defect on MMG and MMTF plates at 37 °C (Fig. ). It was reported that an rlmA deletion mutant was hypersensitive to calcofluor white and a high concentration of Congo red (150 μg/mL), but not to micafungin.Citation4,32) In agreement with these results, ΔrlmA-1 exhibited a growth defect on a YG plate containing calcofluor white, but not a YG plate containing 100 μg/mL Congo red and micafungin (data not shown). ΔrlmA-1 was hypersensitive to Congo red on an MMG plate and was slightly sensitive to calcofluor white and Congo red on an MMTF plate, but it was not sensitive to heat stress on either an MMG plate or an MMTF plate (Fig. ). R429AΔrlmA-1 also exhibited a slight growth defect and sensitivity to Congo red but not to calcofluor white, micafungin, or heat stress compared to R429ApyroA-1 under the pkcA(R429A)-repressing condition (Fig. ). However, under the pkcA(R429A)-inducing condition, it was slightly resistant to calcofluor white and Congo red compared to R429ApyroA-1 (Fig. ).
Table 2. Sequences similar to the consensus Rlm1p binding sequence in the promoter regions of the cell wall-related genes of A. nidulans.
Fig. 4. Stress sensitivities of ΔrlmA-1 and R429AΔrlmA-1.
Notes: 103 conidia of the wild-type strain (A1149/pyroA), ΔrlmA-1, R429ApyroA-1, and R429AΔrlmA-1 were inoculated on MMG, MMTF, and MMG and MMTF containing calcofluor white (CFW), Congo red (CR), or micafungin, and incubated for 72 h at the indicated temperature.
When ΔrlmA-1 and R429AΔrlmA-1 were grown under the pkcA(R429A)-repressing condition and then shifted to the pkcA(R429A)-inducing condition, the mRNA levels of chsB, chsD, csmA, and csmB in R429AΔrlmA-1 were similar to those in ΔrlmA-1 even 1 h after the shift, suggesting that PkcA increases the expression of chsB, chsD, csmA, and csmB through RlmA (Fig. ). In addition, the mRNA levels of chsC and agsB in R429A-1 were 2.86- and 9.32-fold higher, respectively, than those in the wild-type strain 1 h after the shift to the pkcA(R429A)-inducing condition, while those in R429AΔrlmA-1 were 1.75- and 5.19-fold higher, respectively, than those in ΔrlmA-1. This suggests that PkcA increases the expression of chsC and agsB through an RlmA-dependent and RlmA-independent manner. In contrast, the mRNA level of chsG in R429AΔrlmA-1 was much lower than that in ΔrlmA-1, suggesting that under the pkcA(R429A)-inducing condition, chsG repression is RlmA independent.
Transcriptional regulation of cell wall-related genes by PkcA under heat stress
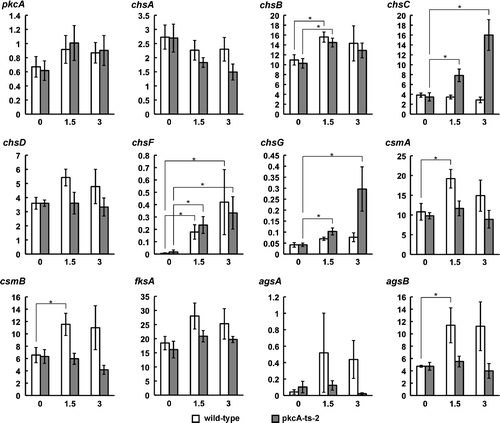
Since we previously showed that PkcA is activated by heat stress,Citation24) PkcA is likely to be involved in the transcriptional regulation of cell wall-related genes under heat stress. When a pkcA temperature-sensitive mutant, pkcA-ts-2,Citation24) was grown at the permissive temperature and then shifted to the restrictive temperature, the mRNA levels of chsB, chsF, csmA, csmB, and agsB in the wild-type strain increased 1.5 h after the shift, while those of csmA, csmB, and agsB in pkcA-ts-2 did not change after the shift (Fig. ). In addition, although the mRNA levels of chsC and chsG in the wild-type strain did not change after the shift, they did increase in pkcA-ts-2 after the shift (Fig. ). These results suggest that PkcA is involved in the transcriptional upregulation of csmA, csmB, and agsB and the downregulation of chsC and chsG under heat stress.
Fig. 5. mRNA levels of the cell wall-related genes under heat stress.
Notes: Total RNAs were prepared from the wild-type strain (A1149) and pkcA-ts-2, which were grown on MMG for 20 h at a permissive temperature (30 °C) and then incubated for the indicated time at a restriction temperature (42 °C). The x-axes indicate the incubation time at 42 °C. The y-axes indicate the relative mRNA levels to that of H2B (%). Data are shown as means ± S.E.M. of four independent experiments. Statistical significance of data relative to the result obtained at 0 h is given by the p-value (*p < 0.05).
Discussion
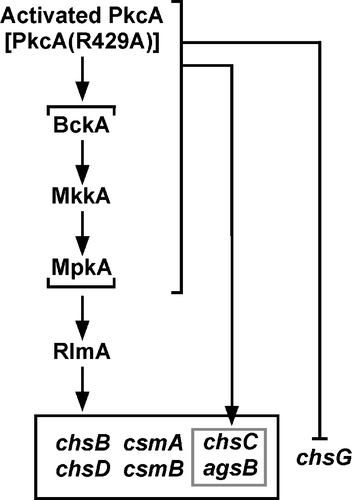
Although PkcA is suggested to play a role in the CWI pathway, it is not known whether PkcA is involved in the transcriptional regulation of cell wall-related genes. In this study, we constructed a strain R429A-1 that expresses pkcA(R429A) under the control of the alcA promoter. In this strain, the mRNA levels of most chitin synthase genes and an α-1,3-glucan synthase gene, agsB, were increased under the pkcA(R429A)-inducing condition; however, these increases were not observed in the rlmA deletion mutant. In addition, the increases in the mRNA levels of csmA, csmB, and agsB observed in the wild-type strain under heat stress were not observed in the pkcA temperature-sensitive mutant, pkcA-ts-2. A model of the signal transduction pathway that regulates the transcription of these cell wall-related genes under the pkcA(R429A)-inducing condition is shown in Fig. .
Fig. 6. A model of the signal transduction pathway that regulates the transcription of the cell wall-related genes under the pkcA(R429A)-inducing condition.
Notes: The signal transduction pathway suggested in this study is shown in this figure. Involvement of BckA, MkkA, and MpkA in the regulation of cell wall-related genes transcription is not investigated in this study and these genes are shown in parenthesis. Details are described in the text.
It was reported that the over expression of pkcA led to increased sensitivity to Congo red and micafungin.Citation18) In this study, we showed that under the pkcA(R429A)-inducing condition, R429A-1 was sensitive to calcofluor white and Congo red, but was resistant to micafungin (Fig. ). The level of MpkA phosphorylation in R429A-1 was higher than that in alcA(p)-pkcA-3 under the pkcA(R429A) and pkcA-inducing condition (Fig. E). Therefore, these differences in the sensitivities to calcofluor white and micafungin between alcA(p)-pkcA-3 and R429A-1 may be caused by variations in PkcA activity. Since the sensitivity of R429A-1 to calcofluor white and Congo red under the pkcA(R429A)-inducing condition was partially suppressed by deletion of rlmA (Fig. ), this sensitivity may be due in part to hyperactivation of RlmA.
The mRNA levels of chsB, chsC, chsD, csmA, csmB, and agsB increased under the pkcA(R429A)-inducing condition, and these increases were dependent on RlmA (Fig. ), suggesting that PkcA upregulates the transcription of these genes through RlmA and probably the MAP kinase cascade in the CWI pathway (Fig. ). In addition, sequences with similarity to the consensus Rlm1p-binding sequence are present in the promoter regions of these genes (Table ).Citation12) These findings suggest that RlmA directly regulates the transcription of these genes. As described in the Introduction, whether MpkA and RlmA are involved in the transcriptional regulation of cell wall-related genes in A. nidulans is a matter of debate. In S. cerevisiae, a small GTPase, Rho1p, functions upstream of Pkc1p in the CWI pathway. Pkc1p binds to Rho1p via its cystine-rich (C1) and homology region 1 (HR1) domains, and activation of the MAP kinase cascade in the CWI pathway is regulated by the interaction between Rho1p and the C1 domain, but not the HR1 domain of Pkc1p.Citation33–35) Thus, it is possible that signals generated by stimuli other than micafungin treatment are transmitted to Pkc1p through Rho1p binding to the HR1 domain and that the signaling pathway activated by pkcA(R429A) overexpression is, at least in part, different from that activated by micafungin treatment.
The mRNA levels of chsC and agsB were increased even in the rlmA deletion mutant under the pkcA(R429A)-inducing condition (Fig. ), suggesting that PkcA also upregulates the expression of these genes in an RlmA-independent manner (Fig. ). In S. cerevisiae, Mpk1p activates a transcription regulatory complex, the SBF complex, which is composed of Swi4p and Swi6p.Citation16) Answi4 and Answi6 were identified as the A. nidulans orthologs of SWI4 and SWI6, respectively.Citation4) However, the consensus sequence recognized by the S. cerevisiae SBF complex is not present in the promoter regions of chsC and agsB.Citation32) Therefore, it is possible that PkcA induces the expression of these genes via an unidentified mechanism.
Although the phosphorylation level of MpkA in R429A-1 was similar to that in the wild-type strain under the pkcA(429A)-repressing condition (Fig. (D)), the conidiation efficiency of R429A-1 was approximately half of that of the wild-type strain, and the mRNA level of chsF in R429A-1 was higher than that in the wild-type stain (Fig. ). These changes may be caused by a slight change in PkcA activity that does not affect the phosphorylation level of MpkA.
Although ΔrlmA-1 was hypersensitive to Congo red on the MMG plate, its sensitivity was reduced on the MMTF plate (Fig. ). Since the mRNA level of chsG was significantly increased when ΔrlmA-1 grown in the MMG medium was shifted to the MMT medium (Fig. ), it is possible that chsG plays a role in the reduction of the sensitivity to Congo red of ΔrlmA-1 on the MMTF plate.
The mRNA levels of csmA, csmB, and agsB in the wild-type strain increased under heat stress; however, these increases were not observed in pkcA-ts-2 (Fig. ), suggesting that PkcA is involved in the transcriptional regulation of these genes under heat stress. Since the rlmA deletion mutant was not hypersensitive to heat stress (Fig. ), RlmA does not appear to play a critical role under heat stress. Since the consensus sequences recognized by Msn2p and Msn4p, which are transcription factors involved in the response to several stresses in S. cerevisiae, are present in the promoter region between csmA and csmB, and that of agsB (Table ),Citation12) it is possible that under heat stress, PkcA regulates these genes through MsnA, an ortholog of Msn2p.Citation36) Under heat stress, the mRNA levels of chsC and chsG in the wild-type strain did not change, whereas the levels did increase in pkcA-ts-2 (Fig. ), suggesting that PkcA is required for the repression of these genes under heat stress. It has been suggested that the expression of AfchsA, which is an A. fumigatus ortholog of chsC, is regulated by the Ca2+ signaling pathway.Citation37,38) In addition, a predicted consensus sequences recognized by CrzA, (G(T/G)GGC(T/A)G(T/G)G), a transcription factor in the Ca2+ signaling pathway in A. nidulans,Citation39) are present in the promoter regions of chsC and chsG (Table ). Many genes that are known to be regulated by CrzA are upregulated in pkcA-ts-2 at the restrictive temperature (Katayama et al., manuscript in preparation). Therefore, under heat stress, the expression of chsC and chsG would be induced by the activation of CrzA in pkcA-ts-2.
Table 3. Sequences similar to the consensus Msn2p and Msn4p binding sequence in the promoter regions of the cell wall-related genes of A. nidulans.
Table 4. Sequences similar to the consensus CrzA binding sequence in the promoter regions of the chsC and chsG genes of A. nidulans.
The results obtained in this study suggest that PkcA is involved in the transcriptional regulation of most chitin synthase genes and agsB. Since the chsB deletion mutant grew very slowly and formed tiny colonies and chsB localized mainly at hyphal tips, chsB was thought to play a crucial role in growth.Citation7–9) chsD is suggested to play compensatory roles for chsB.Citation10) The chsA and chsC double deletion mutant was hypersensitive to cell wall-perturbing agents, suggesting that chsA and chsC are required for CWI.Citation13) The csmA deletion mutant was hypersensitive to cell wall-perturbing agents,Citation29) and the expression of csmA was increased in the chsA and chsC double deletion mutant,Citation40) suggesting that CsmA plays a role in cell wall repair. Both CsmA and CsmB have a myosin motor-like domain at their N-termini and were shown to have similar compensatory functions in growth.Citation12,29,41–43) csmB is located adjacent to csmA in a head-to-head configuration on the genome, and the expression pattern of csmB was similar to that of csmA (Fig. and ).Citation11,12) Thus, it is probable that CsmB is also involved in cell wall repair. An increase in α-1,3-glucan content was observed in A. fumigatus when the chitin synthase genes were deleted,Citation44) and the expression of the α-1,3-glucan synthase gene was induced under cell wall stress in A. nidulans and Aspergillus niger.Citation4,45) These results suggest that α-glucan functions as a compensatory component of the cell wall. Taken together, PkcA likely plays a critical role in maintaining CWI by exquisitely regulating the expression of chsB, chsC, chsD, csmA, csmB, and agsB.
Supplemental material
The supplemental material for this paper is available at http://dx.doi.org/10.1080/09168451.2014.973365.
Supplemental Materials
Download Zip (602.4 KB)Acknowledgements
We thank Aya Ono-Tanaka for construction of the plasmid, pUCpyroA2. This work was done using the facilities of the Biotechnology Research Center of the University of Tokyo.
Additional information
Funding
Notes
Abbreviations: CWI, cell wall integrity; PKC, protein kinase C; MAP kinase, mitogen-activated protein kinase.
References
- Latgé JP. The cell wall: a carbohydrate armour for the fungal cell. Mol. Microbiol. 2007;66:279–290.10.1111/mmi.2007.66.issue-2
- de Groot PW, Brandt BW, Horiuchi H, Ram AF, de Koster CG, Klis FM. Comprehensive genomic analysis of cell wall genes in Aspergillus nidulans. Fungal Genet. Biol. 2009;46:S72–S81.10.1016/j.fgb.2008.07.022
- Horiuchi H. Functional diversity of chitin synthases of Aspergillus nidulans in hyphal growth, conidiophore development and septum formation. Med. Mycol. 2009;47:S47–S52.10.1080/13693780802213332
- Fujioka T, Mizutani O, Furukawa K, Sato N, Yoshimi A, Yamagata Y, Nakajima T, Abe K. MpkA-dependent and -independent cell wall integrity signaling in Aspergillus nidulans. Eukaryot. Cell. 2009;6:1497–1510.
- Yoshimi A, Sano M, Inaba A, Kokubun Y, Fujioka T, Mizutani O, Hagiwara D, Fujikawa T, Nishimura M, Yano S, Kasahara S, Shimizu K, Yamaguchi M, Kawakami K, Abe K. Functional analysis of the α-1,3-glucan synthase genes agsA and agsB in Aspergillus nidulans: AgsB is the major α-1,3-glucan synthase in this fungus. PLoS ONE. 2013;8:e54893.10.1371/journal.pone.0054893
- Firon A, Beauvais A, Latgé JP, Couvé E, Grosjean-Cournoyer MC, D’Enfert C. Characterization of essential genes by parasexual genetics in the human fungal pathogen Aspergillus fumigatus: impact of genomic rearrangements associated with electroporation of DNA. Genetics. 2002;161:1077–1087.
- Yanai K, Kojima N, Takaya N, Horiuchi H, Ohta A, Takagi M. Isolation and characterization of two chitin synthase genes from Aspergillus nidulans. Biosci. Biotechnol. Biochem. 1994;58:1828–1835.10.1271/bbb.58.1828
- Borgia PT, Iartchouk N, Riggle PJ, Winter KR, Koltin Y, Bulawa CE. The chsB gene of Aspergillus nidulans is necessary for normal hyphal growth and development. Fungal Genet. Biol. 1996;20:193–203.10.1006/fgbi.1996.0035
- Fukuda K, Yamada K, Deoka K, Yamashita S, Ohta A, Horiuchi H. Class III chitin synthase chsB of Aspergillus nidulans localizes at the sites of polarized cell wall synthesis and is required for conidial development. Eukaryot. Cell. 2009;8:945–956.10.1128/EC.00326-08
- Ichinomiya M, Motoyama T, Fujiwara M, Takagi M, Horiuchi H, Ohta A. Repression of chsB expression reveals the functional importance of class IV chitin synthase gene chsD in hyphal growth and conidiation of Aspergillus nidulans. Microbiology. 2002;148:1335–1347.
- Takeshita N, Ohta A, Horiuchi H. csmA, a gene encoding a class V chitin synthase with a myosin motor-like domain of Aspergillus nidulans, is translated as a single polypeptide and regulated in response to osmotic conditions. Biochem. Biophys. Res. Commun. 2002;298:103–109.10.1016/S0006-291X(02)02418-X
- Takeshita N, Yamashita S, Ohta A, Horiuchi H. Aspergillus nidulans class V and VI chitin synthases CsmA and CsmB, each with a myosin motor-like domain, perform compensatory functions that are essential for hyphal tip growth. Mol. Microbiol. 2006;59:1380–1394.10.1111/j.1365-2958.2006.05030.x
- Fujiwara M, Ichinomiya M, Motoyama T, Horiuchi H, Ohta A, Takagi M. Evidence that the Aspergillus nidulans class I and II chitin synthase genes, chsC and chsA, share critical roles in hyphal wall integrity and conidiophore development. J. Biochem. 2000;127:359–366.10.1093/oxfordjournals.jbchem.a022616
- Ichinomiya M, Yamada E, Yamashita S, Ohta A, Horiuchi H. Class I and class II chitin synthases are involved in septum formation in the filamentous fungus Aspergillus nidulans. Eukaryot. Cell. 2005;4:1125–1136.10.1128/EC.4.6.1125-1136.2005
- Levin DE. Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2005;69:262–291.10.1128/MMBR.69.2.262-291.2005
- Levin DE. Regulation of cell wall biogenesis in Saccharomyces cerevisiae: the cell wall integrity signaling pathway. Genetics. 2011;189:1145–1175.10.1534/genetics.111.128264
- Jung US, Levin DE. Genome-wide analysis of gene expression regulated by the yeast cell wall integrity signalling pathway. Mol. Microbiol. 1999;34:1049–1057.10.1046/j.1365-2958.1999.01667.x
- Ichinomiya M, Uchida H, Koshi Y, Ohta A, Horiuchi H. A protein kinase C-encoding gene, pkcA, is essential to the viability of the filamentous fungus Aspergillus nidulans. Biosci. Biotechnol. Biochem. 2007;71:2787–2799.10.1271/bbb.70409
- Ronen R, Sharon H, Levdansky E, Romano J, Shadkchan Y, Osherov N. The Aspergillus nidulans pkcA gene is involved in polarized growth, morphogenesis and maintenance of cell wall integrity. Curr. Genet. 2007;51:321–329.10.1007/s00294-007-0129-y
- Teepe AG, Loprete DM, He Z, Hoggard TA, Hill TW. The protein kinase C orthologue PkcA plays a role in cell wall integrity and polarized growth in Aspergillus nidulans. Fungal Genet. Biol. 2007;44:554–562.10.1016/j.fgb.2006.10.001
- Herrmann M, Spröte P, Brakhage AA. Protein kinase C (PkcA) of Aspergillus nidulans is involved in penicillin production. Appl. Environ. Microbiol. 2006;72:2957–2970.10.1128/AEM.72.4.2957-2970.2006
- Chellegatti MA, Yuvamoto PD, Said S. Role of phospholipase C and protein kinase C in Aspergillus nidulans during growth on pectin or glucose: effects on germination and duplication cycle. Folia Microbiol. 2010;55:228–232.10.1007/s12223-010-0033-6
- Colabardini AC, de Castro PA, de Gouvêa PF, Savoldi M, Malavazi I, Goldman MH, Goldman GH. Involvement of the Aspergillus nidulans protein kinase C with farnesol tolerance is related to the unfolded protein response. Mol. Microbiol. 2010;78:1259–1279.10.1111/j.1365-2958.2010.07403.x
- Katayama T, Uchida H, Ohta A, Horiuchi H. Involvement of protein kinase C in the suppression of apoptosis and in polarity establishment in Aspergillus nidulans under conditions of heat stress. PLoS ONE. 2012;7:e50503.10.1371/journal.pone.0050503
- Futagami T, Seto K, Kajiwara Y, Takashita H, Omori T, Takegawa K, Goto M. The putative stress sensor protein MtlA is required for conidia formation, cell wall stress tolerance, and cell wall integrity in Aspergillus nidulans. Biosci. Biotechnol. Biochem. 2014;78:326–335.10.1080/09168451.2014.878218
- Rowlands RT, Turner G. Nuclear and extranuclear inheritance of oligomycin resistance in Aspergillus nidulans. Mol. Gen. Genet. 1973;126:201–216.10.1007/BF00267531
- Yu JH, Hamari Z, Han KH, Seo JA, Reyes-Domínguez Y, Scazzocchio C. Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet. Biol. 2004;41:973–981.10.1016/j.fgb.2004.08.001
- Oakley CE, Weil CF, Kretz PL, Oakley BR. Cloning of the riboB locus of Aspergillus nidulans. Gene. 1987;53:293–298.10.1016/0378-1119(87)90019-9
- Horiuchi H, Fujiwara M, Yamashita S, Ohta A, Takagi M. Proliferation of intrahyphal hyphae caused by disruption of csmA, which encodes a class V chitin synthase with a myosin motor-like domain in Aspergillus nidulans. J. Bacteriol. 1999;181:3721–3729.
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685.10.1038/227680a0
- Watanabe M, Chen CY, Levin DE. Saccharomyces cerevisiae PKC1 encodes a protein kinase C (PKC) homolog with a substrate specificity similar to that of mammalian PKC. J. Biol. Chem. 1994;269:16829–16836.
- Kovács Z, Szarka M, Kovács S, Boczonádi I, Emri T, Abe K, Pócsi I, Pusztahelyi T. Effect of cell wall integrity stress and RlmA transcription factor on asexual development and autolysis in Aspergillus nidulans. Fungal Genet. Biol. 2013;54:1–14.10.1016/j.fgb.2013.02.004
- Nonaka H, Tanaka K, Hirano H, Fujiwara T, Kohno H, Umikawa M, Mino A, Takai Y. A downstream target of RHO1 small GTP-binding protein is PKC1, a homolog of protein kinase C, which leads to activation of the MAP kinase cascade in Saccharomyces cerevisiae. EMBO J. 1995;14:5931–5938.
- Schmitz HP, Jöckel J, Block C, Heinisch JJ. Domain shuffling as a tool for investigation of protein function: substitution of the cysteine-rich region of Raf kinase and PKC η for that of yeast Pkc1p. J. Mol. Biol. 2001;311:1–7.10.1006/jmbi.2001.4848
- Schmitz HP, Heinisch JJ. Evolution, biochemistry and genetics of protein kinase C in fungi. Curr. Genet. 2003;43:245–254.10.1007/s00294-003-0403-6
- Han KH, Prade RA. Osmotic stress-coupled maintenance of polar growth in Aspergillus nidulans. Mol. Microbiol. 2002;43:1065–1078.10.1046/j.1365-2958.2002.02774.x
- Fortwendel JR, Juvvadi PR, Perfect BZ, Rogg LE, Perfect JR, Steinbach WJ. Transcriptional regulation of chitin synthases by calcineurin controls paradoxical growth of Aspergillus fumigatus in response to caspofungin. Antimicrob. Agents Chemother. 2010;54:1555–1563.10.1128/AAC.00854-09
- Pinchai N, Perfect BZ, Juvvadi PR, Fortwendel JR, Cramer RA Jr, Asfaw YG, Heitman J, Perfect JR, Steinbach WJ. Aspergillus fumigatus calcipressin cbpa is involved in hyphal growth and calcium homeostasis. Eukaryot. Cell. 2009;8:511–519.10.1128/EC.00336-08
- Hagiwara D, Kondo A, Fujioka T, Abe K. Functional analysis of C2H2 zinc finger transcription factor CrzA involved in calcium signaling in Aspergillus nidulans. Curr. Genet. 2008;54:325–338.10.1007/s00294-008-0220-z
- Yamada E, Ichinomiya M, Ohta A, Horiuchi H. The class V chitin synthase gene csmA is crucial for the growth of the chsA chsC double mutant in Aspergillus nidulans. Biosci. Biotechnol. Biochem. 2005;69:87–97.10.1271/bbb.69.87
- Takeshita N, Ohta A, Horiuchi H. CsmA, a class V chitin synthase with a myosin motor-like domain, is localized through direct interaction with the actin cytoskeleton in Aspergillus nidulans. Mol. Biol. Cell. 2005;16:1961–1970.10.1091/mbc.E04-09-0761
- Tsuizaki M, Takeshita N, Ohta A, Horiuchi H. Myosin motor-like domain of the class VI chitin synthase CsmB is essential to its functions in Aspergillus nidulans. Biosci. Biotechnol. Biochem. 2009;73:1163–1167.10.1271/bbb.90074
- Tsuizaki M, Ohta A, Horiuchi H. Myosin motor-like domain of class VI chitin synthase CsmB of Aspergillus nidulans is not functionally equivalent to that of class V chitin synthase CsmA. Biosci. Biotechnol. Biochem. 2013;77:369–374.
- Mellado E, Dubreucq G, Mol P, Sarfati J, Paris S, Diaquin M, Holden DW, Rodriguez-Tudela JL, Latgé JP. Cell wall biogenesis in a double chitin synthase mutant (chsG-/chsE-) of Aspergillus fumigatus. Fungal Genet. Biol. 2003;38:98–109.10.1016/S1087-1845(02)00516-9
- Damveld RA, vanKuyk PA, Arentshorst M, Klis FM, van den Hondel CA, Ram AF. Expression of agsA, one of five 1,3-α-D-glucan synthase-encoding genes in Aspergillus niger, is induced in response to cell wall stress. Fungal Genet. Biol. 2005;42:165–177.10.1016/j.fgb.2004.11.006
