Abstract
The xylanase (Xyn10B) that strongly adsorbs on microcrystalline cellulose was isolated from Driselase. The Xyn10B contains a Carbohydrate-binding module family 1 (CBM1) (IrpCBMXyn10B) at N-terminus. The canonical essential aromatic residues required for cellulose binding were conserved in IrpCBMXyn10B; however, its adsorption ability was markedly higher than that typically observed for the CBM1 of an endoglucanase from Trametes hirsuta (ThCBMEG1). An analysis of the CBM-GFP fusion proteins revealed that the binding capacity to cellulose (7.8 μmol/g) and distribution coefficient (2.0 L/μmol) of IrpCBMXyn10B-GFP were twofold higher than those of ThCBMEG1-GFP (3.4 μmol/g and 1.2 L/μmol, respectively), used as a reference structure. Besides the canonical aromatic residues (W24-Y50-Y51) of typical CBM1-containing proteins, IrpCBMXyn10B had an additional aromatic residue (Y52). The mutation of Y52 to Ser (IrpCBMY52S-GFP) reduced these adsorption parameters to 4.4 μmol/g and 1.5 L/μmol, which were similar to those of ThCBMEG1-GFP. These results indicate that Y52 plays a crucial role in strong cellulose binding.
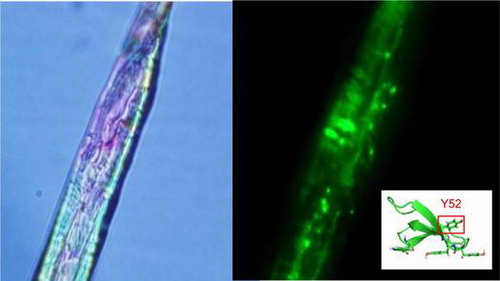
Fluorescence microscopy image of IrpCBMXyn10B-GFP adsorbed on cotton (A: bright field, B: dark field) and three dimensional structure of CBM1 of Xyn10B from Irpex lacteus.
Carbohydrate-binding module family 1 (CBM1) is a cellulose-binding domain that is present primarily in fungal lignocellulose-degrading proteins. CBM1 consists of 30–40 amino acid residues and is connected to a core domain via a linker region. It facilitates the adsorption of proteins on cellulose and enhances the degradation of cellulose in the lignocellulosic complex, especially on the surface.Citation1) Three essential amino acid residues were previously shown to be crucial for binding and are located on the same shelf-like surface. These residues adsorb the glucopyranose rings exposed on the cellulose surface via hydrophobic interactions. The motifs of these three amino acid residues were found to be Y-YY, W-WY, W-YY, Y-WY, and Y-FY, and a correlation has been reported between their hydrophobicity and the adsorptive ability of CBM1. Consequently, the efficiency by which cellulase degrades insoluble cellulose was shown to increase as the capacity for adsorption increased.Citation2,3) In contrast, the absence of CBM1 markedly reduces the degradation of crystalline cellulose due to a decrease in adsorption. As observed in Trichoderma reesei cellobiohydrolase (CBH) type I and type II (named Cel7A and Cel6A, respectively), the adsorption behaviors of their CBM1s vary. The adsorption of Cel7A CBM1 (Y-YY) occurs over a short period, and the protein shifts to another region of cellulose. On the other hand, Cel6A CBM1 (W-YY) adsorbs on cellulose for a longer period than Cel7A because of its higher binding parameters.Citation4,5) New types of CBM1 with the motifs (Y-W-N-Y and Y-W-S-F) have recently been reported in Oomycetes sp.Citation6) They were found to be structurally similar to fungal CBM1, but are associated with domains involved in adhesion to endogenous or exogenous ligands. From this, it seems likely that the function of lignocellulose-degrading (and other) enzymes may be affected by the adsorption characteristics of their CBM1 modules.
CBMs have been categorized into 69 families from their amino acid sequences according to the CAZy (carbohydrate-active enzymes) database.Citation7) Several hundred CBM1s have been identified to date based on amino acid sequence similarities. Although CBM1s are a small domain and the majority is present in fungal cellulolytic enzymes, CBM1s are also distributed in several hemicellulases and non-hydrolytic proteins such as swollenin, polysaccharide monooxygenase, cellobiose dehydrogenase, and carbohydrate esterase.Citation8–13) Although differences in the amino acid sequences of CBM1 should affect its binding ability, this feature has not been functionally analyzed in detail.
Irpex lacteus, a white rot fungus, produces an efficient plant cell wall-degrading system.Citation14) I. lacteus strain MC-2 is used in the production of the commercial enzyme preparation, Driselase. At least 11 cellulolytic and hemicellulolytic enzymes were identified from a draft genomic sequence of I. lacteus strain NK-1 that contains CBM1 domains with a various amino acid motifs for binding cellulose. In this study, the cellulose-binding proteins in Driselase were fractionated on the basis of their differences in their affinity to cellulose.Citation15) A new type of CBM1 (IrpCBMXyn10B) with a unique binding motif (W-YYY) was identified in one of I. lacteus GH10 xylanase. Binding characteristics of this CBM1 were determined using CBM1 fused to a GFP protein.
Materials and methods
Chemicals, strains, and growth conditions
Driselase is a commercial enzyme preparation from Kyowa Hakko Co. (Tokyo, Japan). Microcrystalline cellulose (MC, for column chromatography, Merck KGaA, Darmstadt, Germany) was used as an adsorbent for enzyme fractionation and as a substrate for the binding studies. Cotton used as adsorbent was obtained from Hakujuji Corporation (Tokyo, Japan). A stock culture of I. lacteus strain NK-1 was isolated earlier by Nisizawa.Citation16) Escherichia coli DH5α, purchased from Takara (Shiga, Japan), was used as the host strain for plasmid extraction and was grown at 37 °C in Luria–Bertani medium containing 50 μg/ml ampicillin. Plasmid pBluescript II SK(+) (Agilent Technologies, CA, USA) was used as the subcloning vector. The E. coli Origami B strain (Merck KGaA, Darmstadt, Germany) and plasmid pRSET/EmGFP (Invitrogen, Carlsbad, CA, USA) were used for the expression of the CBM1 fused to green fluorescence protein (CBM-GFPs).
Fractionation of proteins from Driselase
Ten grams of MC was added to 1000 mL of 1% Driselase solution, and this suspension was agitated overnight at 4 °C. MC was then collected by filtration and washed with 600 ml of cold 0.1 M sodium acetate buffer (pH 5.0). Recovered non-adsorbed proteins are assigned as fraction 1. The proteins adsorbed on MC were eluted with 500 mL of 1 mM sodium acetate buffer (pH 5.0) at 4 °C (fraction 2) and at 37 °C (fraction 3). Proteins remaining on MC (fraction 4) were eluted with 2% sodium dodecyl sulfate (SDS) containing 0.06% 2-mercaptoethanol by boiling the mixture for 3 min. Each elute was subjected to SDS polyacrylamide gel electrophoresis (PAGE), and the gel was stained for proteins with Coomassie Brilliant Blue R-250.
Amino acid sequencing
After the SDS-PAGE analysis, the 42 kDa protein was stained and recovered by AE-1310 EzStain Reverse (ATTO, Japan). The 42 kDa protein was incubated at 37 °C for 16 h with 0.25 μg of V8 protease (Wako, Japan) in 50 mM sodium phosphate buffer (pH 7.8). Native and digested proteins were transferred to PVDF membranes (GE Healthcare, UK) after electrophoresis and then subjected to a PPSQ-31 protein sequencer (Shimadzu, Japan). The resulting sequences were collated to the database of the I. lacteus strain NK-1draft genome sequence. Nucleotide sequence data of Xyn10B from I. lacteus strain MC-2 is available in the DDBJ/EMBL/GenBank databases under the accession number AB933639.
Plasmid construction of CBM-GFP fusion proteins
The full-length cDNA of Xyn10B was amplified by PCR (PCR conditions: 95 °C for 5 min, followed by 30 cycles at 98 °C for 10 s, 49 °C for 15 s, 72 °C for 90 s, followed by a final extension step at 72 °C, 5 min) from the first strand cDNA of I. lacteus strain MC-2 using Prime STAR HS DNA polymerase (Takara, Shiga, Japan) with the sense primer (5′-AAGCTTAAGCTCAACCTCACAAGA-3′) and antisense primer (5′-TCTAGATTACGAGAGAGAGCCTGCATGA-3′). The amplified fragment (1166 bp) was ligated to the SmaI site of pBluescript II SK(+) (named pBS-Xyn10B). The CBM1 region of Xyn10B was also amplified from pBS-Xyn10B using Prime STAR HS DNA polymerase (PCR conditions: 95 °C for 5 min, followed by 30 cycles at 98 °C for 10 s, 60 °C for 15 s, 72 °C for 30 s, followed by a final extension step at 72 °C, 5 min) with the sense primer (5′-AGGATCCGCTGTCGCTGAGTGG-3′, BamHI site is underlined) and antisense primer (5′-GCCATGGAGCCAGAGCCGGCGG-3′, NcoI site is underlined). The amplified IrpCBMXyn10B fragment (138 bp) was also ligated to the SmaI site of pBluescript II SK(+). The resulting plasmid was then digested by BamHI and NcoI, and the IrpCBMXyn10B gene was inserted into the same sites of pRSET/EmGFP for fusion with the vector encoding GFP and histidine tag. The resulting plasmid (named pRSET-IrpCBMXyn10B-GFP) was used as an E. coli expression vector. The fusion protein expression vector of the CBM1 derived from Trametes hirsuta endoglucanase 1 (EG1) and GFP was prepared as described by Nozaki et al.Citation17).
CBM1 mutant fusion protein (IrpCBMF42S-GFP, IrpCBMY52S-GFP) expression vectors were constructed according to the manual of the PrimeSTAR Mutagenesis basal kit (Takara, Japan) using mutation primers (5′-TCTCCCTCTGTCTGCACGGTTATCAAC-3′ and 5′-GCAGACAGAGGGAGAGTCACACGTGGT-3′ for IrpCBMF42S-GFP; 5′-TACTACTCTCAGTGCTTGCCCGCCGGC-3′and 5′-GCACTGAGAGTAGTAGCTGTTGATAAC-3′ for IrpCBMY52S-GFP, serine mutation site is underlined) and pRSET-IrpCBMXyn10B-GFP as a template (PCR conditions: 95 °C for 5 min, followed by 30 cycles at 98 °C for 10 s, 52 °C for 15 s, 72 °C for 30 s, followed by a final extension step at 72 °C, 5 min).
The nucleotide sequences of the obtained plasmids were confirmed by sequencing and used for the transformation of E. coli Origami B strain (Novagen, NJ), the expression strain.
Production and purification of CBM-GFPs
E. coli Origami B strains were grown in 100 ml of Luria–Bertani medium containing 100 μg/mL ampicillin with shaking at 180 rpm at 30 °C, induced with 1 mM isopropyl β-D-thiogalactopyranoside (IPTG) after 12 h. After 24 h cultivation at 30 °C, cells were harvested by centrifugation and suspended in 20 mM phosphate buffer (pH 7.4) containing 0.5 M NaCl. Cells were disrupted by an ultrasonic treatment, and each CBM-GFP was purified from the supernatant using a His-trap FF column (GE Healthcare, UK) according to the manual. Purified CBM-GFPs were confirmed by SDS-PAGE as a single protein band.
Binding study of CBM-GFP fusion proteins
The reaction mixture (1 mL) consisted of 0.02% MC and various amounts of protein in 10 mM sodium acetate buffer (pH 5.0). The reaction mixture was incubated at 4 °C for 2 h with rotation at 12 rpm. After adsorption, the reaction mixture was centrifuged for 2 min at 13,000 rpm. Non-adsorbed CBM-GFPs in the supernatant were subjected to SDS-PAGE. The cellulose with adsorbed CBM-GFPs were washed twice with 0.1 mL of deionized water and suspended in a total volume of 1 mL. The proteins were eluted and also subjected to SDS-PAGE by same method of the fraction 4 from Driselase (described above).
The amount of adsorbed protein was measured by determining the intensity of fluorescence (excitation/emission = 487/509 nm) in the supernatant of the reaction mixture. Binding parameters were determined by fitting the data to the Langmuir equation.
In this equation, [B] and [F] are the concentrations of bound and free CBM-GFPs (mol per gram of cellulose), respectively. Amax and Kad are the values of binding capacity (μmol/g of cellulose) and the distribution coefficient (μM−1) of bound protein, respectively. The Amax and Kad values in this equation can be calculated by single reciprocal plots ([B]/[F] vs. [F]).Citation18)
Fluorescent images of cotton fibers with CBM-GFP proteins
The CBM-GFP fusion protein (1 μM) was incubated with 1 mg of cotton in 10 mM sodium acetate buffer (pH 5.0) in a total volume of 1 mL. After 2 h at 4 °C with rotation at 12 rpm, the cotton was washed twice with 1 mL of the same buffer. The adsorbed CBM-GFP fusion protein was observed using the BX-51 fluorescence microscope (Olympus, Japan).
Results
Fractionation of proteins from Driselase
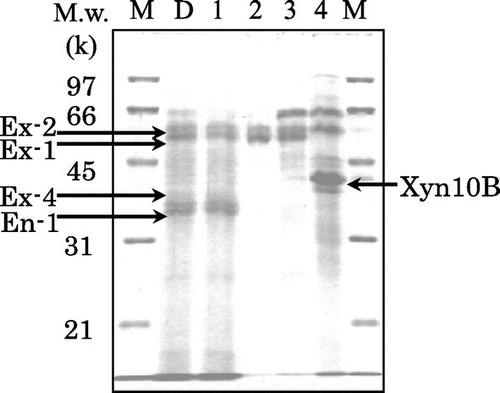
Proteins from Driselase were separated into four fractions using a cellulose affinity batch chromatography. The fractions were analyzed by SDS-PAGE (Fig. ). Several bands were identified as exo- and endo-type cellulases on the basis of earlier data were purified and characterized from Driselase by Kanda. et al.Citation19,20). A considerable portion of endoglucanase I and CBH II (designated as En-1 and Ex-4, 35 and 39 kDa, respectively) that lost their CBM1s due to proteolysis did not adsorb to cellulose.Citation21,22) The enzymes that were trapped on cellulose by means of their CBM1s were CBH Ia (designated as Ex-1, 53 kDa) and CBH Ib (designated as Ex-2, 55 kDa). Both CBH Is were identified as isoforms with different glycosylations.Citation23) Proteins of our main interest were those adsorbed firmly on cellulose, not eluted from cellulose even with a solution of 1 M NaCl containing 1% (v/v) Triton X-100, or 0.06% 2-mercaptoethanol (data not shown). These strongly adsorbed proteins were released from cellulose as fraction 4, after 3 min boiling the cellulose suspension in a solution of 2% SDS, containing 0.06% 2-mercaptoethanol. The most abundant protein in fraction 4 (mol. mass 42 kDa, Fig. ) was subjected to amino acid sequencing.
Fig. 1. SDS-PAGE of proteins fractionated from Driselase on the basis of adsorbing ability on microcrystalline cellulose.
Amino acid sequence of the 42 kDa protein
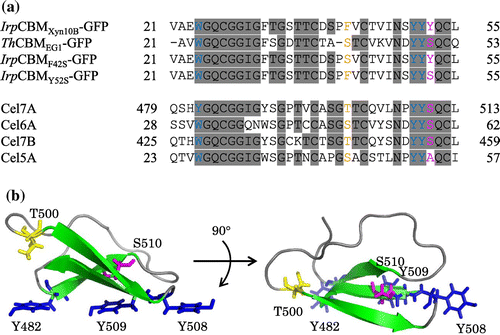
The N-terminal sequence of the 42 kDa protein from fraction 4 was determined to be VAEWGQXGGIGFT (X was unidentified). The internal amino acid sequences of this protein were also determined to be NSMKWDATENTRGQFTFSG and IFNEDGTFRSSVFYNVL for the two peptide fragments with masses of 10 and 22 kDa, respectively. These were identical to the sequence of CBM1 and a part of the catalytic domain of GH10 xylanase (Xyn10B) in Driselase. The deduced amino acid sequence from the cDNA of Xyn10B consisted of 383 amino acid residues, of which the first 20 residues represented the signal sequence. The apparent molecular mass (42 kDa) was approximately 4 kDa higher than that calculated from genetic information (38 kDa). A search of the Genbank database revealed that the Xyn10B catalytic domain shares sequence similarities with those of the GH10 xylanases, from Phanerochaete carnosa HHB-10118-sp (83%, Genbank accession No. EKM58934), Thermoascus aurantiacus xylanase (63%, 2BNJ_A), and T. reesei xylanase III (56%, BAA89465.2). Of these, Xyn10B and P. carnosa xylanases had CBM1s. A comparison of the binding motif of IrpCBMXyn10B to other reported CBM1s from T. reesei revealed that IrpCBMXyn10B contained two additional aromatic residues (F42 and Y52) in the conserved amino acid sequence (Fig. (a)). The essential residues of IrpCBMXyn10B for cellulose binding were considered to be W24, Y50, and Y51 (corresponding to Y482, Y508, and Y509 of CBM1 from T. reesei Cel7A shown in Fig. (b)), and the additional aromatic residue, Y52, was located on different site from the flat surface consisting of three aromatic residues that formed part of the binding motif. The residue corresponding to F42 and Y52 in Xyn10B is replaced by serine or threonine in well-characterized CBM1s, such as that of EG1 from T. hirsuta, and Cel7A, Cel6A, and Cel7B from T. reesei.
Fig. 2. Amino acid sequences and three-dimensional structures of CBM1s.
Binding study of CBM-GFPs on cellulose
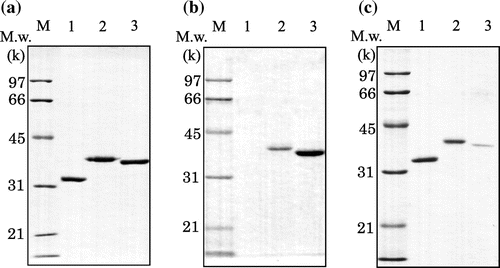
To clarify the strong adsorption of Xyn10B on crystalline cellulose, only the CBM1 region of Xyn10B was expressed as a fusion protein with GFP (IrpCBMXyn10B-GFP), as shown in Fig. (a). The fluorescence of the construct enabled easy quantification of the adsorption. An additional fusion protein between GFP and CBM1 from EG1 (designated as ThCBMEG1-GFP) was also constructed for comparative purposes. The adsorption behavior of IrpCBMXyn10B-GFP on cellulose should be similar to that of Xyn10B, as both proteins had the same CBM1 that had higher adsorption than ThCBMEG1-GFP (Fig. (b) and (c)). This result suggested that the strong adsorption of Xyn10B should be attributed to CBM1 and not to the catalytic domain.
Fig. 3. SDS-PAGE analysis of CBM-GFPs.
Adsorption isotherms were also determined according to the one-site Langmuir model (Fig. ). The Amax and Kad values of each CBM-GFP fusion protein are shown in Table . The Amax (7.8 μmol/g) and Kad (2.0 l/μmol) values of IrpCBMXyn10B-GFP were approximately twofold higher than those of ThCBMEG1-GFP (3.4 μmol/g and 1.2 L/μmol, respectively), even though both proteins had the same binding motif (W-YY). The isotherms of ThCBMEG1-GFP were almost the same as those reported in the CBM1s from T. reesei Cel7A and Cel5A.Citation24) The major difference between earlier characterized CBM1s and the IrpCBMXyn10B is the presence of additional amino acid residues, F42 and Y52. To investigate the role of these two amino acid residues, mutant proteins, IrpCBMF42S-GFP and IrpCBMY52S-GFP, were constructed and their adsorption properties compared with the original structure. The Amax and Kad values of IrpCBMF42S-GFP (7.9 μmol/g and 1.8 L/μmol, respectively) were similar to that of IrpCBMXyn10B-GFP, while the adsorption ability of IrpCBMY52S-GFP (4.4 μmol/g and 1.5 L/μmol, respectively) was reduced significantly, up to the values observed for ThCBMEG1-GFP. These results serve as strong evidence that the aromatic residue Y52, of the unique cellulose-binding module motif (W-YYY), plays an important role in the high binding affinity of IrpCBMXyn10B.
Fig. 4. Adsorption isotherms of wild-type and mutated CBM-GFPs to microcrystalline cellulose.
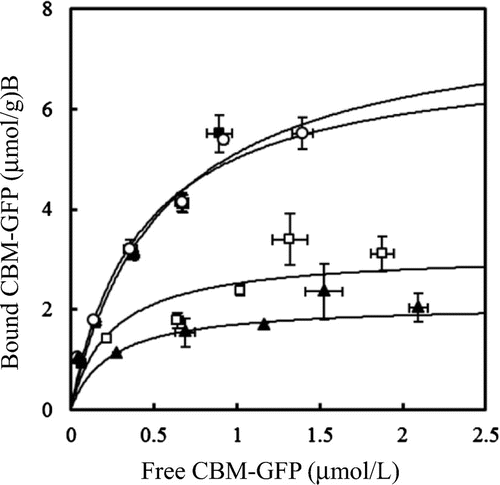
Table 1. Adsorption parameters of wild-type and mutated CBM-GFPs on microcrystalline cellulose from various sources.
Microscopic images of the adsorption of CBM-GFP on cellulose
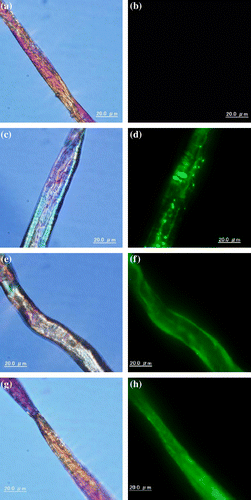
CBM-GFPs adsorbed on cotton were examined under a fluorescence microscope (Fig. ). GFP without CBM1 was not adsorbed on cotton; therefore, no fluorescence was observed on cellulose (Fig. (b)). ThCBMEG1-GFP (Fig. (f)) and the mutant protein, IrpCBMY52S-GFP (Fig. (h)), were adsorbed on cotton surface uniformly, as the fluorescence was observed the whole surface of cotton cellulose. On the other hand, the fluorescence of IrpCBMXyn10B-GFP (Fig. (d)) was detected in a form of fluorescent patches. This observation suggests that IrpCBMXyn10B-GFP molecules adsorb on cellulose as molecular aggregates formed due to hydrophobic intermolecular interaction.
Fig. 5. Bright-field (left) and fluorescent detection (right) image of GFP (a,b), IrpCBMXyn10B-GFP (c, d), ThCBMEG1-GFP (e, f), and IrpCBMY52S-GFP (g, h) adsorbed on cotton.
One can assume that the distinct adsorption behaviors of the native and mutant CBM1s may result from the difference in their three-dimensional structures.
Discussion
The adsorption of enzymes on cellulose is an important step for its effective degradation. The role of CBM1 in efficiency of hydrolysis is believed to be the enhancement of the concentration of the enzyme molecules on cellulose surface.Citation2) In the present study, fractionation of proteins from the commercial enzyme preparation Driselase based on different affinities to cellulose (Fig. ) led to isolation of a 42 kDa xylanase, which exhibited irreversible adsorption on MC. The enzyme could not be desorbed from cellulose without heating with a detergent such as SDS. Sugimoto et al.Citation15) reported that more than 80% of cellulases with CBM1s were eluted from a cellulose affinity column under mild conditions. There are examples of cellulose-binding domains that adsorb onto cellulose irreversibly under physiological conditions. One is the CBM1 of Cel6A from T. reesei which has an additional S–S bond making the structure more rigid than the CBM1 of Cel7A.Citation5) Another strongly bound CBM is the CBM2a of xylanase 10A from Cellulomonas fimi.Citation25–27) The CBM2a was shown to have a more planar binding face.
CBM1 of I. lacteus Xyn10B (IrpCBMXyn10B), present at the N-terminal region of the enzyme (amino acids 21–55), also irreversibly binds to cellulose. This property is obviously determined by its architecture and number of aromatic amino acids interacting with cellulose surface. Unique features of IrpCBMXyn10B, in comparison with CBM1s of other enzymes, are two additional aromatic residues, F42 and Y52 (replaced to serine, threonine, or alanine in CBM1s of EG1 and other cellulases from T. reesei), and three contiguous aromatic residues (Y50, Y51, and Y52). EG1 and other cellulases from T. reesei (Fig. (A)) which do not show such a strong affinity for cellulose as IrpCBMXyn10B contain serine at the place of Y52. We have clearly demonstrated that replacement of Y52 by serine in IrpCBMXyn10B resulted in considerable decrease in affinity for cellulose. The adsorption isotherms of IrpCBMY52S-GFP were significantly lower than those of IrpCBMxyn10B-GFP (Fig. ). The adsorption parameters of IrpCBMY52S-GFP closely matched those of ThCBMEG1-GFP, constructed here for comparative purposes (Table ). To clarify possible role of F42 in the adsorption, a mutant CBMF42S was prepared and fused with GFP. The adsorption parameters of the construct IrpCBMF42S-GFP were similar to those of the native IrpCBMXyn10B-GFP which speaks for the fact that F42 is not involved in the adsorption to cellulose.
Table 2. Distribution of CBM1s with the W-YYY motif in glycoside hydrolase family enzymes and other proteins related to cellulose degradation in the CAZy database (http://www.cazy.org/).
The strong adsorption to cellulose of the unique IrpCBMXyn10B could be seen well under a fluorescence microscope. An unexpected observation was that IrpCBMXyn10B-GFP molecule did not adsorb on cotton evenly. Fluorescence was concentrated in green fluorescent in spots which suggests that the construct adsorbs on cellulose in an aggregated form. The aggregation of IrpCBMXyn10B-GFP was observed in solution also in the absence of cellulose and could be enhanced by increasing the protein and salt concentration (Supplemental Fig. 1). On the other hand, IrpCBMY52S-GFP molecule adsorbed on cotton evenly and did not show signs of aggregation in solution under similar conditions. These results pointed out that additional tyrosine Y52 plays an important role not only in the adsorption process but also in the aggregation of IrpCBMXyn10B-GFP molecules. A possibility cannot be excluded that Y52 is not involved directly in hydrophobic interaction of flat face consisting of three aromatic amino acids with cellulose surface, but is responsible for formation of CBM1 aggregates. Relevant to the above considerations is the three-dimensional structure of CBM1 of Cel7A from T. reesei determined by NMR spectroscopy by Mattinen et al.Citation28). (Fig. (b)). The structure shows that the side chain of S510 (corresponded to Y52 of IrpCBMXyn10B) of CBM1 from Cel7A faces to opposite side of cellulose-binding flat face which is consisted of Y482, Y508, and Y509. The homology modeling of the IrpCBMXyn10B using the Swiss model (http://swissmodel.expasy.org/) suggested that the aromatic side chain of Y52 may also be not located on the flat surface for cellulose binding, but it might be involved in the change of hydrophobicity of the environment on the rough face of IrpCBMXyn10B. It is noteworthy that the IrpCBMXyn10B-GFP shows the remarkable aggregation and still maintains the binding ability. Linder et al.Citation29) reported that a “CBM1 hetero-construct” prepared by fusion of the N-terminal CBM1 of Cel6A and C-terminal of CBM1 of Cel7A also showed higher affinity for cellulose than single domains, and it is suggested that the aggregation of IrpCBMXyn10B domains can make the multiple binding face and this is seemed to be one of the possibility for strong adsorption.
On the CAZy database (http://www.cazy.org/), many CBM1s which have W-YYY motif have been found in hemicellulases such as xylanase and mannanase (19/55) (Table ). On the other hand, only 5 CBM1s which have W-YYY motif were found in CBH-type cellulases though most of all CBH have at least one CBM1. As the adsorption ability of CBM1s which have W-YYY motif on cellulose is thought to be remarkably strong, this property may not be favorable for processive manner of degradation. A weaker adsorption of enzymes on cellulose should be more convenient in this regard. Processive cellulose degradation and reducing sugar production from filter paper were in fact reduced by introduction of additional CBM1 to the hydrolase.Citation30) On the other hand, this property can be suitable for hemicellulases such as xylanase and mannanase because of the close association of hemicellulose with cellulose.Citation31–33) Strong adsorption of enzymes on cellulose can be considered the natural economic way of keeping them continuously working on plant biomass. We should also note that the picture of the role of CBMs in microbial degradation of plant cell wall polysaccharides became more complicated after recent report on possible role in binding the enzymes to cellulose by highly glycosylated linkers.Citation34)
Supplemental material
The supplemental material for this paper is available at http://dx.doi.org/10.1080/09168451.2014.996203.
Supplemental Materials
Download Zip (138.1 KB)Acknowledgments
This study was performed under the Development of Technology for High Efficiency Bioenergy Conversion Project supported by New Energy and Industrial Technology Development Organization (NEDO) and also supported by a Grant-in-Aid for Global COE Program by Ministry of Education, Culture, Sports, Science, and Technology.
Additional information
Funding
Notes
Abbreviations: CBM1, carbohydrate-binding module family 1; CBD, cellulose-binding domain; GH, glycoside hydrolase family; MC, microcrystalline cellulose; CBH, cellobiohydrolase; EG, endoglucanase; GFP, green fluorescent protein.
References
- Lehtio J, Sugiyama J, Gustavsson M, Fransson L, Linder M, Teeri TT. The binding specificity and affinity determinants of family 1 and family 3 cellulose binding modules. Proc. Nat. Acad. Sci. USA. 2003;100:484–489.10.1073/pnas.212651999
- Thongekkaew J, Ikeda H, Masaki K, Iefuji H. Fusion of cellulose binding domain from Trichoderma reesei CBHI to Cryptococcus sp. S-2 cellulase enhances its binding affinity and its cellulolytic activity to insoluble cellulosic substrates. Enzyme Microb. Technol. 2013;52:241–246.10.1016/j.enzmictec.2013.02.002
- Hamada N, Kodaira R, Nogawa M, Shinji K, Ito R, Amano Y, Shimosaka M, Kanda T, Okazaki M. Role of cellulose-binding domain of exocellulase I from White rot basidiomycete Irpex lacteus. J. Biosci. Bioeng. 2001;91:359–362.10.1016/S1389-1723(01)80151-1
- Linder M, Teeri TT. The cellulose-binding domain of the major cellobiohydrolase of Trichoderma reesei exhibits true reversibility and a high exchange rate on crystalline cellulose. Proc. Nat. Acad. Sci. USA. 1996;93:12251–12255.10.1073/pnas.93.22.12251
- Carrard G, Linder M. Widely different off rates of two closely related cellulose-binding domains from Trichoderma reesei. Eur. J. Biochem. 1999;262:637–643.10.1046/j.1432-1327.1999.00455.x
- Larroque M, Barriot R, Bottin A, Barre A, Rougé P, Dumas B, Gaulin E. The unique architecture and function of cellulose-interacting proteins in oomycetes revealed by genomic and structural analyses. BMC Genomics. 2012;13:605.10.1186/1471-2164-13-605
- Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The carbohydrate-active enzymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 2009;37:D233–D238.10.1093/nar/gkn663
- Khan SN, Akter MZ, Sims PFG. Cloning and expression analysis of two endo-1,4-β-xylanase genes from Phanerochaete chrysosporium. Plant Tissue Cult. 2002;12:57–68.
- Tenkanen M, Buchert J, Viikari L. Binding of hemicellulases on isolated polysaccharide substrates. Enzyme Microb. Technol. 1995;17:499–505.10.1016/0141-0229(94)00050-2
- Saloheimo M, Paloheimo M, Hakola S, Pere J, Swanson B, Nyyssönen E, Bhatia A, Ward M, Penttilä M. Swollenin, a Trichoderma reesei protein with sequence similarity to the plant expansins, exhibits disruption activity on cellulosic materials. Eur. J. Biochem. 2002;269:4202–4211.10.1046/j.1432-1033.2002.03095.x
- Horn SJ, Vaaje-Kolstad G, Westereng B, Eijsink VG. Novel enzymes for the degradation of cellulose. Biotechnol. Biofuels. 2012; doi:10.1186/1754-6834-5-45.
- Igarashi K, Verhagen MFJM, Samejima M, Schulein M, Eriksson K-EL, Nishino T. Cellobiose Dehydrogenase from the Fungi Phanerochaete chrysosporium and Humicola insolens: a flavohemoprotein from Humicola insolens contains 6-hydroxy-fad as the dominant active cofactor. J. Biol. Chem. 1999;274:3338–3344.10.1074/jbc.274.6.3338
- Wong DW, Chan VJ, McCormack AA, Hirsch J, Biely P. Functional cloning and expression of the Schizophyllum commune glucuronoyl esterase gene and characterization of the recombinant enzyme. Biotechnol. Res. Int. 2012;2012: 951267. doi:10.1155/2012/951267.
- Novotný Č, Cajthaml T, Svobodová K, Šušla M, Šašek V. Irpex lacteus, a white-rot fungus with biotechnological potential — review. Folia Microbiol. 2009;54:375–390.10.1007/s12223-009-0053-2
- Sugimoto N, Igarashi K, Samejima M. Cellulose affinity purification of fusion proteins tagged with fungal family 1 cellulose-binding domain. Protein Expression and Purif. 2012;82:290–296.10.1016/j.pep.2012.01.007
- Nisizawa K, Hashimoto Y. Cellulose-splitting enzymes. VI. Difference in the specificities of cellulase and β-glucosidase from Irpex lacteus. Arch. Biochem. Biophys. 1959;81:211–222.10.1016/0003-9861(59)90191-2
- Nozaki K, Nishijima H, Arai T, Mizuno M, Sato N, Amano Y. Regulation of adsorption behavior of carbohydrate-binding module family 1 and endo-β-1,4-glucanase onto crystalline cellulose. J. Appl. Glycosci. 2011;58:133–138.10.5458/jag.jag.JAG-2011_007
- Tomme P, Boraston A, McLean B, Kormos J, Creagh AL, Sturch K, Gilkes NR, Haynes CA, Warren RAJ, Kilburn DG. Characterization and affinity applications of cellulose-binding domains. J. Chromatogr. B. Biomed. Sci. Appl. 1998;715:283–296.10.1016/S0378-4347(98)00053-X
- Kanda T, Nakakubo S, Wakabayashi K, Nisizawa K. Purification and properties of exo-cellulase of avicelase type from a wood-rotting fungus, Irpex lacteus (Polyporus tulipiferae). J. Biochem. 1978;84:1217–1226.
- Kanda T, Wakabayashi K, Nisizawa K. Purification and properties of a lower-molecular-weight endo-cellulase from Irpex lacteus (Polyporus tulipiferae). J. Biochem. 1980;87:1625–1634.
- Toda H, Nagahata N, Amano Y, Nozaki K, Kanda T, Okazaki M, Shimosaka M. Gene cloning of cellobiohydrolase II from the white rot fungus Irpex lacteus MC-2 and its expression in Pichia pastoris. Biosci. Biotechnol., Biochem. 2008;72:3142–3147.10.1271/bbb.80316
- Toda H, Takada S, Oda M, Amano Y, Kanda T, Okazaki M, Shimosaka M. Gene cloning of an endoglucanase from the basidiomycete Irpex lacteus and Its cDNA expression in Saccharomyces cerevisiae. Biosci. Biotechnol., Biochem. 2005;69:1262–1269.10.1271/bbb.69.1262
- Hamada N, Ishikawa K, Fuse N, Kodaira R, Shimosaka M, Amano Y, Kanda T, Okazaki M. Purification, characterization and gene analysis of exo-cellulase II (Ex-2) from the white rot basidiomycete Irpex lacteus. J. Biosci. Bioeng. 1999;87:442–451.10.1016/S1389-1723(99)80092-9
- Medve J, Karlsson J, Lee D, Tjerneld F. Hydrolysis of microcrystalline cellulose by cellobiohydrolase I and endoglucanase II from Trichoderma reesei: adsorption, sugar production pattern, and synergism of the enzymes. Biotechnol. Bioeng. 1998;59:621–634.10.1002/(ISSN)1097-0290
- Creagh AL, Ong E, Jervis E, Kilburn DG, Haynes CA. Binding of the cellulose-binding domain of exoglucanase Cex from Cellulomonas fimi to insoluble microcrystalline cellulose is entropically driven. Proc. Nat. Acad. Sci. USA. 1996;93:12229–12234.10.1073/pnas.93.22.12229
- Jervis EJ, Haynes CA, Kilburn DG. Surface diffusion of cellulases and their isolated binding domains on cellulose. J. Biol. Chem. 1997;272:24016–24023.10.1074/jbc.272.38.24016
- McLean BW, Bray MR, Boraston AB, Gilkes NR, Haynes CA, Kilburn DG. Analysis of binding of the family 2a carbohydrate-binding module from Cellulomonas fimi xylanase 10A to cellulose: specificity and identification of functionally important amino acid residues. Protein Eng. 2000;13:801–809.10.1093/protein/13.11.801
- Mattinen ML, Kontteli M, Kerovuo J, Linder M, Annila A, Lindeberg G, Reinikainen T, Drakenberg T. Three-dimensional structures of three engineered cellulose-binding domains of cellobiohydrolase I from Trichoderma reesei. Protein Sci. 1997;6:294–303.
- Linder M, Salovuori I, Ruohonen L, Teeri TT. Characterization of a double cellulose-binding domain. Synergistic high affinity binding to crystalline cellulose. J. Biol. Chem. 1996;271:21628–21272.
- Zheng F, Ding S. Processivity and enzymatic mode of a glycoside hydrolase family 5 endoglucanase from Volvariella volvacea. Appl. Environ. Microbiol. 2013;79:989–996.10.1128/AEM.02725-12
- Gill J, Rixon JE, Bolam DN, Mcqueen-Mason SJ, Simpson PJ, Williamson MP, Hazlewood GP, Gilbert HJ, Creagh AL, Ong E, Jervise E, Kilburn DG, Haynes CA. The type II and X cellulose-binding domains of Pseudomonas xylanase A potentiate catalytic activity against complex substrates by a common mechanism. Biochem. J. 1999;342:473–480.10.1042/0264-6021:3420473
- Black GW, Rixon JE, Clarke JH, Hazlewood GP, Theodorou MK, Morris P, Gilbert HJ. Evidence that linker sequences and cellulose-binding domains enhance the activity of hemicellulases against complex substrates. Biochem. J. 1996;319:515–520.
- Hägglund P, Eriksson T, Collén A, Nerinckx W, Claeyssens M, Stålbrand H. A cellulose-binding module of the Trichoderma reesei β-mannanase Man5A increases the mannan-hydrolysis of complex substrates. J. Biotechnol. 2003;101:37–48.10.1016/S0168-1656(02)00290-0
- Payne CM, Resch MG, Chen L, Crowley MF, Himmel ME, Taylor LE II, Sandgren M, Stahlberg J, Stals I, Tan Z, Beckham GT. Glycosylated linkers in multimodular lignocellulose-degrading enzymes dynamically bind to cellulose. Proc. Nat. Acad. Sci. USA. 2013;110:14646–14651.10.1073/pnas.1309106110
- Medve J, Ståhlberg J, Tjerneld F. Isotherms for adsorption of cellobiohydrolase I and II from Trichoderma reesei on microcrystalline cellulose. Appl. Biochem. Biotechnol. 1997;66:39–56.10.1007/BF02788806
- Georgelis N, Yennawar NH, Cosgrove DJ. Structural basis for entropy-driven cellulose binding by a type-A cellulose-binding module (CBM) and bacterial expansin. Proc. Nat. Acad. Sci. USA. 2012;109:14830–14835.10.1073/pnas.1213200109
