Abstract
Koji mold, Aspergillus oryzae, has been used for the production of sake, miso, and soy sauce for more than one thousand years in Japan. Due to the importance, A. oryzae has been designated as the national micro-organism of Japan (Koku-kin). A. oryzae has been intensively studied in the past century, with most investigations focusing on breeding techniques and developing methods for Koji making for sake brewing. However, the understanding of fundamental biology of A. oryzae remains relatively limited compared with the yeast Saccharomyces cerevisiae. Therefore, we have focused on studying the cell biology including live cell imaging of organelles, protein vesicular trafficking, autophagy, and Woronin body functions using the available genomic information. In this review, I describe essential findings of cell biology of A. oryzae obtained in our study for a quarter of century. Understanding of the basic biology will be critical for not its biotechnological application, but also for an understanding of the fundamental biology of other filamentous fungi.
Graphical abstract
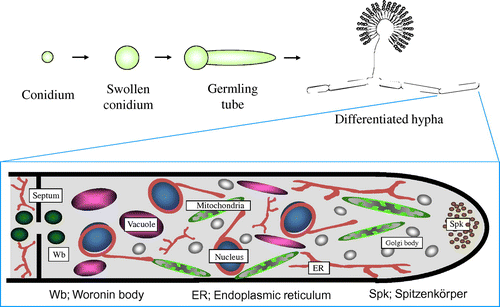
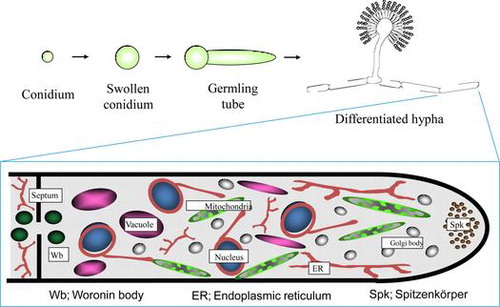
Schematic model of Aspergillus oryzae organelles, which was drawn based on the results of visualization by expression of EGFP fusion proteins.
Koji mold, Aspergillus oryzae, has been used for the production of sake, miso, and soy sauce for more than ten centuries in Japan. Due to the importance of these foods in Japanese culture, A. oryzae has been designated as the national micro-organism of Japan (Koku-kin) by the Brewing Society of Japan in 2006.Citation1)
A. oryzae has been intensively studied in the past century, with most investigations focusing on breeding techniques and developing methods for Koji production for sake brewing. However, the understanding of fundamental biological processes of A. oryzae remains relatively limited when compared with the yeast Saccharomyces cerevisiae. The main reasons for this lack of progress are that A. oryzae is an imperfect fungus without a sexual life cycle and is not amenable to classical genetic manipulation procedures. Therefore, our group was interested in studying the cell biology of A. oryzae using the available genomic information for this organism.
I. Genome analysis of A. oryzae
In 1994, the electrophoretic karyotype of A. oryzae was determined by transverse altering-field electrophoresis. Seven chromosomal DNA bands with estimated sizes of 7.0, 5.2, 5.0, 4.5, 4.0, 3.7, and 2.8 Mb were identified using 13 cloned genes, including ribosomal DNA, as probes. The intensity of ethidium staining and the results of Southern blotting with 100 random clones isolated from A. oryzae suggested that the smallest band migrated as doublet and that the total genome size was approximately 35 Mb.Citation2)
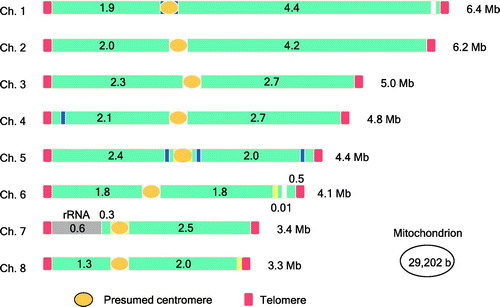
In 1997, the A. oryzae Expressed Sequence Tag (EST) analysis project team performed comprehensive analyses of the mRNAs expressed in A. oryzae under various culture conditions, including nutrient-rich and carbon-starved liquid cultures, liquid-germinated conidia, solid medium with alkaline pH, and solid-state wheat bran and rice cultures.Citation3) At the completion of the EST project in 2001, the A. oryzae Genome Consortium, which was comprised of industry, university, and government partners, determined that the 37-Mb genome of A. oryzae contains 12,074 genes on 8 chromosomes and is expanded by 7–9 Mb in comparison with the genomes of Aspergillus nidulans and Aspergillus fumigatus (Fig. ).Citation4,5) The findings from these consortia on the genome structure of A. oryzae greatly advanced molecular biological studies of this fungus, for which numerous papers have now been published, predominantly by Japanese research groups.
Fig. 1. Schematic drawing of the A. oryzae genome.Citation4)
II. Visualization of A. oryzae organelles using fluorescent proteins (EGFP and DsRed)
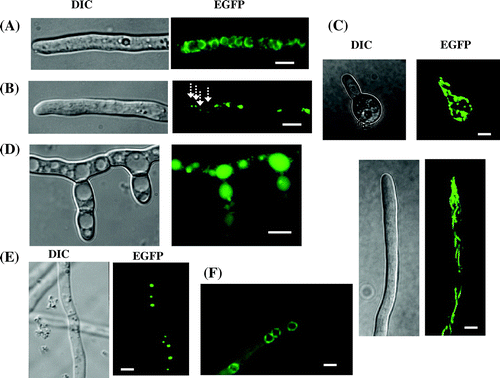
When we began studying the molecular biology of Koji mold about 25 years ago, the working model for the cellular structure of filamentous fungi, including A. oryzae, was based mainly on that of yeast due to a lack of detailed microscopic observation. To improve the understanding of the fine structure of living cells of A. oryzae, our laboratory attempted to perform live cell imaging of all organelles using the available genomic information to construct enhanced green fluorescence protein (EGFP) fusion constructs. To date, we have successfully visualized all cellular organelles of A. oryzae, including the endoplasmic reticulum,Citation6–8) Golgi apparatus,Citation7) mitochondria,Citation9) vacuoles,Citation10) peroxisomes,Citation11) nucleus,Citation12–14) Woronin body,Citation15) Spitzenkörper,Citation16)and septum (Fig. ).Citation16,17)
Fig. 2. Visualization of organelles in A. oryzae by expression of EGFP fusion proteins. (A) Endoplasmic reticulum,Citation7) (B) Golgi apparatus,Citation7) (C) mitochondria,Citation9) (D) vacuoles,Citation10) (E), peroxisomes,Citation11) and (F), nuclei.
We have also established a technique for the labeling of different organelles with EGFP and red fluorescence protein (DsRed) to allow the simultaneous visualization of target components involved in various cellular processes, such as vesicular trafficking, endocytosis,Citation18) and autophagy,Citation19) as described in the following sections. Based on the results of these analyses, we determined the structure of the A. oryzae cell (Fig. ).
III. Analysis of protein vesicular trafficking in A. oryzae cells
A. oryzae secretes abundant quantities of α-amylase and has one of the highest secretion abilities among eukaryotes. However, understanding of vesicular trafficking in filamentous fungi is relatively limited from a molecular biological standpoint. Hence, to visualize the secretory process of α-amylase in A. oryzae cells we performed localization analysis using amyB-EGFP in liquid and solid media. As shown in Fig. , this analysis revealed that a filamentous fungus-specific organelle, called the Spitzenkörper, is present at the hyphal tip region and is involved in the accumulation of secretion vesicles containing secretory enzymes. The amyB-EGFP fluorescence signal is more intense on agar plates compared to liquid culture, implying amyB is secreted at higher levels from cells on solid medium.Citation16)
Fig. 4. Localization of α-amylase fused with EGFP in liquid and agar cultures.Citation16).
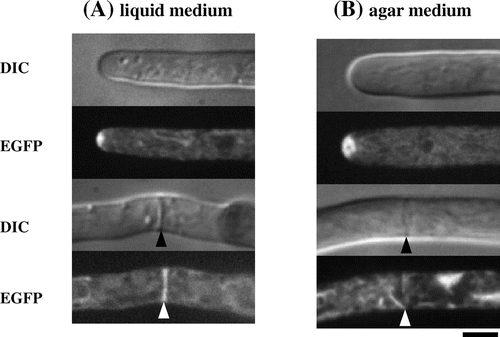
Filamentous fungi such as A. oryzae are multicellular organisms consisting of numerous thin cells sectioned by partitions into hyphae, which exhibit polarized growth that proceeds through extension of highly metabolically active hyphal tip cells. To analyze secretion-related organelles, we performed systematic localization analysis using EGFP and the approximately 20 SNARE proteins to decide specificity in membrane fusion of the secretion vesicles.Citation7) Using this approach, we determined that the secretion pathway in A. oryzae proceeds from the endoplasmic reticulum via Golgi apparatus and the Spitzenkörper to the cell membrane.Citation20) In addition, we successfully visualized the process of endocytosis, which has been tightly coupled with protein secretion (exocytosis), for the first time in a filamentous fungus.Citation18,21) In A. oryzae, the endosome moves at high speed (approximately 4–5 μm/s) along microtubules, as can be seen in the Supplemental movie of our paper published in Biochem Biophys Res Commun.Citation21)
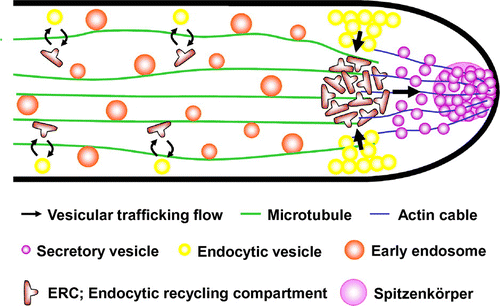
In A. oryzae, secretion vesicles are moved by a long-distance transport system comprised of microtubules to the apical tip, where the vesicles are transported by an actin cable to the membrane for secretion of their contents. Fluorescent microscopic observation revealed that v-SNARE on the secretion vesicle is endocytosed immediately after fusion with the cell membrane, and is then recycled for incorporation into new secretion vesicles. This recycling process appears to be critical for sustaining high secretion levels by A. oryzae. The proposed model for the endocytic recycling of v-SNARE in the apical tip of A. oryzae is shown in Fig. .Citation22)
Fig. 5. Proposed model for the endocytic recycling in apical tip supporting high protein secretion of A. oryzae.Citation22)
IV. Analysis of vacuolar function and autophagy in A. oryzae
Vacuoles are lytic compartments with acidic pH that have important roles in metabolite storage and the homeostasis of cytosolic ions and pH. In filamentous fungi, small spherical vacuoles are abundantly present in apical regions, whereas tubular vacuoles form a continuum in subapical regions, and large, developed spherical vacuoles are found in basal regions. Based on the observed distribution of vacuoles, it appears that they play distinctive roles in filamentous fungi depending on the mycelial region. To observe vacuoles in greater detail, we constructed strains of A. oryzae that express AoVam3p, a vacuolar t-SNARE, tagged with EGFP. The fusion protein successfully labeled to the membrane of pleiomorphic vacuoles, including tubular structures (Fig. ).Citation23)
Fig. 6. Pleiomorphic vacuoles in A. oryzae.Citation23)
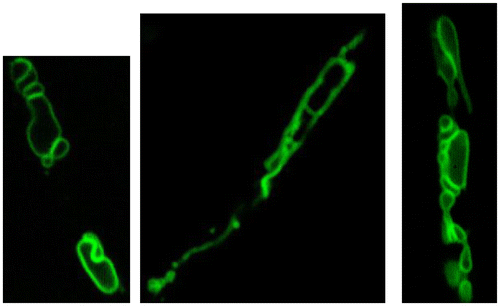
A. oryzae forms colonies by the elongation of apical cells and also extends aerial hyphae towards the air. By functional analysis of vacuole-related genes, we determined that tubular vacuoles develop in both aerial hyphae and hyphae located in the central regions of colonies, suggesting that tubular vacuoles carry nutrients from basal hyphae in contact with the underlying solid nutrient medium.
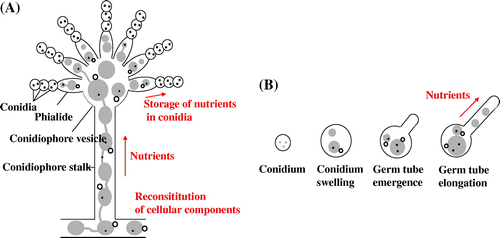
Autophagy is a conserved degradation process of eukaryotes that mediates the incorporation of cytoplasmic proteins and organelles into vacuoles for bulk degradation, which allows recycling of the cytoplasmic components as nutrients to support cell survival.Citation24) Such nutrient recycling is likely critical for the survival of multicellular fungi, which are capable of translocating nutrients along hyphae in order to maximize proliferation at the population level.Citation25) In fact, it has been shown in A. oryzae that cytoplasmic organelles, including mitochondria and peroxisomes, of older hyphae located in the colony center undergo autophagy-mediated degradation. Several lines of evidence suggest that nutrients obtained through such degradation processes are translocated to active mycelial regions with higher demand for nutrients, such as growing hyphal fronts and aerial hyphae.Citation26) In addition, autophagy-deficient mutants of filamentous fungi are typically incapable of forming aerial hyphae, which are not in direct contact with nutrients, further supporting the role of autophagy-mediated degradation and translocation of the resulting nutrients to support non-assimilating hyphae. Studies in A. oryzae using several Aoatg knockout mutants have revealed that autophagy is necessary for aerial hyphae formation, conidiation, and conidial germination (Fig. ).Citation19,27–29)
Fig. 7. Schematic diagram for the putative flow of nutrients recycled by autophagy during conidiation and germination.Citation27)
Recently, we discovered that A. oryzae is capable of degrading entire nuclei by autophagy under starvation conditions, which is the first report of such a process in filamentous fungi.Citation30) Notably, this has not been observed in mononuclear organisms, including yeast, and therefore appears to be a unique ability of A. oryzae, which contains many nuclei in each cell. Although the primary function of the nucleus is generally considered to be the expression of genes encoded in the chromosomal DNA, the nucleus also serves as a nutrient source when cells experience starvation in A. oryzae.
V. Functional analysis of the Woronin body, a filamentous fungi-specific organelle
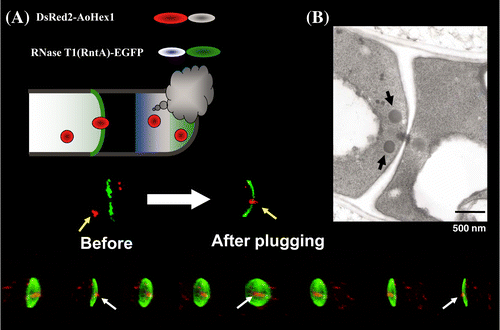
A. oryzae hyphae consist of numerous thin cells that are partitioned by septa. A pore located in the center of the septum allows for cytoplasmic streaming between cells. Although this allows for communication between cells, adjacent cells are susceptible to injury or death when even one cell is damaged. To prevent such injury, a specific organelle called the Woronin body exists near the septum in filamentous fungi, including A. oryzae, and serves to plug the septal pore in response to cell damage (Fig. ).Citation11,15,31) We cloned the Aohex1 gene, which encodes a protein constituting the Woronin body, and determined that the AoHex1 multimer is formed in peroxisomes and plays an essential role in survival against catastrophic cell injury.Citation32) It is important to note that the septal pore is not constitutively open and that fluctuations between the open–closed state is dependent on the behavior of the AoSO protein in response to environmental stress.Citation33,34)
Fig. 8. Dual fluorescent images of an A. oryzae strain expressing DsRed2–AoHex1 and RNase T1–EGFP fusion proteins.Citation15)
The functional analysis of Woronin body brought a new aspect of biotin biosynthesis. Biotin is an essential cofactor involved in a number of carboxylation and decarboxylation reactions. Among eukaryotes, only plants and many species of fungi are able to synthesize biotin. Although the final steps of biotin biosynthesis occur in mitochondria, the cellular location, and components involved in the initial events are poorly understood. Recently, analysis of peroxisome-deficient mutants of A. oryzae, which were constructed for functional analysis of the Woronin body, revealed that first steps of biotin biosynthesis occur in the peroxisome.Citation35) This represents a novel role for peroxisomes in not only the filamentous fungus A. oryzae, but also in the plant Arabidopsis thaliana.
VI. Conclusion and future perspectives
The cell biology of filamentous fungi has been mainly studied using two model lab strains, Aspergillus nidulans and Neurospora crassa. However, the industrially important fungus A. oryzae is now amenable to such studies after the completion of its whole genome sequence. The findings from these studies are contributing to practical applications, such as heterologous protein production by autophagy-deficient mutants.Citation36)
In Japan, there is an old saying of “Onko-chishin,” which roughly translates to “Consult the past if you want to learn about the future.” Since Koji mold, A. oryzae, has been used for more than one thousand years for the production of sake, miso, and soy sauce, understanding of the cell biology of A. oryzae will be critical for not its biotechnological application, but also for an understanding of the fundamental biology of other filamentous fungi.
Acknowledgments
I would like to thank all collaborators whose works are published in the original papers cited here. This review was supported by a Grant-in-Aid for Scientific Research (B) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Disclosure statement
No potential conflict of interest was reported by the author.
Additional information
Funding
Notes
This review was written in response to the author’s receipt of the Japan Society for Bioscience, Biotechnology, and Agrochemistry Senior Scientists Award in 2011.
References
- Ichishima E. Unique enzymes of Aspergillus fungi used in Japanese bioindustries. Nova Science: Novinka; 2012.
- Kitamoto K, Kimura K, Gomi K, Kumagai C. Electrophoretic karyoptype and gene assignment to chromosome of Aspergillus oryzae. Biochemistry. 1994;58:1467–1470.
- Akao T, Sano M, Yamada O, Akeno T, Fujii K, Goto K, Ohashi-Kunihiro S, Takase K, Yasukawa-Watanabe M, Yamaguchi K, Kurihara Y, Maruyama J, Juvvadi PR, Tanaka A, Hata Y, Koyama Y, Yamaguchi S, Kitamoto N, Gomi K, Abe K, Takeuchi M, Kobayashi T, Horiuchi H, Kitamoto K, Kashiwagi Y, Machida M, Akita O. Analysis of expressed sequence tags from the fungus Aspergillus oryzae cultured under different conditions. DNA Res. 2007;14:47–57.10.1093/dnares/dsm008
- Machida M, Asai K, Sano M, Tanaka T, Kumagai T, Terai G, Kusumoto K, Arima T, Akita O, Kashiwagi Y, Abe K, Gomi K, Horiuchi H, Kitamoto K, Kobayashi T, Takeuchi M, Denning DW, Galagan JE, Nierman WC, Yu J, Archer DB, Bennett JW, Bhatnagar D, Cleveland TE, Fedorova ND, Gotoh O, Horikawa H, Hosoyama A, Ichinomiya M, Igarashi R, Iwashita K, Juvvadi PR, Kato M, Kato Y, Kin T, Kokubun A, Maeda H, Maeyama N, Maruyama J, Nagasaki H, Nakajima T, Oda K, Okada K, Paulsen I, Sakamoto K, Sawano T, Takahashi M, Takase K, Terabayashi Y, Wortman JR, Yamada O, Yamagata Y, Anazawa H, Hata Y, Koide Y, Komori T, Koyama Y, Minetoki T, Suharnan S, Tanaka A, Isono K, Kuhara S, Ogasawara N, Kikuchi H. Genome sequencing and analysis of Aspergillus oryzae. Nature. 2005;438:1157–1161.10.1038/nature04300
- Kobayashi T, Abe K, Asai K, Gomi K, Juvvadi PR, Kato M, Kitamoto K, Takeuchi M, Machida M. Genomics of Aspergillus oryzae. Biosci. Biotechnol., Biochem. 2007;71:646–670.10.1271/bbb.60550
- Maruyama J, Kikuchi S, Kitamoto K. Differential distribution of the endoplasmic reticulum network as visualized by the BipA–EGFP fusion protein in hyphal compartments across the septum of the filamentous fungus, Aspergillus oryzae. Fungal Genet. Biol. 2006;43:642–654.10.1016/j.fgb.2005.11.007
- Kuratsu M, Taura A, Shoji JY, Kikuchi S, Arioka M, Kitamoto K. Systematic analysis of SNARE localization in the filamentous fungus Aspergillus oryzae. Fungal Genet. Biol. 2007;44:1310–1323.10.1016/j.fgb.2007.04.012
- Maruyama J, Kitamoto K. Differential distribution of the endoplasmic reticulum network in filamentous fungi. FEMS Microbiol. Lett. 2007;272:1–7.10.1111/fml.2007.272.issue-1
- Mabashi Y1, Kikuma T, Maruyama J, Arioka M, Kitamoto K. Development of a versatile expression plasmid construction system for Aspergillus oryzae and its application to visualization of mitochondria. Biosci. Biotechnol., Biochem. 2006;70:1882–1889.10.1271/bbb.60052
- Ohneda M, Arioka M, Nakajima H, Kitamoto K. Visualization of vacuoles in Aspergillus oryzae by expression of CPY–EGFP. Fungal Genet. Biol. 2002;37:29–38.10.1016/S1087-1845(02)00033-6
- Escano CS, Juvvadi PR, Jin FJ, Takahashi T, Koyama Y, Yamashita S, Maruyama J, Kitamoto K. Disruption of the Aopex11-1 gene involved in peroxisome proliferation leads to impaired Woronin body formation in Aspergillus oryzae. Eukaryotic Cell. 2009;8:296–305.10.1128/EC.00197-08
- Maruyama J, Nakajima H, Kitamoto K. Visualization of nuclei in Aspergillus oryzae with EGFP and analysis of the number of nuclei in each conidium by FACS. Biosci. Biotechnol., Biochem. 2001;65:1504–1510.10.1271/bbb.65.1504
- Maruyama J, Nakajima H, Kitamoto K. Observation of EGFP-visualized nuclei and distribution of vacuoles in Aspergillus oryzae arpA null mutant. FEMS Microbiol. Lett. 2002;206:57–61.10.1111/fml.2002.206.issue-1
- Ishi K, Maruyama J, Juvvadi PR, Nakajima H, Kitamoto K. Visualizing nuclear migration during conidiophore development in Aspergillus nidulans and Aspergillus oryzae: multinucleation of conidia occurs through direct migration of plural nuclei from phialides and confers greater viability and early Germination in Aspergillus oryzae. Biosci. Biotechnol., Biochem. 2005;69:747–754.10.1271/bbb.69.747
- Maruyama J, Juvvadi PR, Ishi K, Kitamoto K. Three-dimensional image analysis of plugging at the septal pore by Woronin body during hypotonic shock inducing hyphal tip bursting in the filamentous fungus Aspergillus oryzae. Biochem. Biophys. Res. Commun. 2005;331:1081–1088.10.1016/j.bbrc.2005.03.233
- Hayakawa Y1, Ishikawa E, Shoji JY, Nakano H, Kitamoto K. Septum-directed secretion in the filamentous fungus Aspergillus oryzae. Mol. Microbiol. 2011;81:40–55.10.1111/mmi.2011.81.issue-1
- Masai K, Maruyama J, Nakajima H, Kitamoto K. In vivo visualization of the distribution of a secretory protein in Aspergillus oryzae hyphae using the RntA-EGFP fusion protein. Biosci. Biotechnol., Biochem. 2003;67:455–459.10.1271/bbb.67.455
- Higuchi Y, Shoji JY, Arioka M, Kitamoto K. Endocytosis is crucial for cell polarity and apical membrane recycling in the filamentous fungus Aspergillus oryzae. Eukaryotic Cell. 2009;8:37–46.10.1128/EC.00207-08
- Kikuma T, Ohneda M, Arioka M, Kitamoto K. Functional analysis of the ATG8 homologue Aoatg8 and role of autophagy in differentiation and germination in Aspergillus oryzae. Eukaryotic Cell. 2006;5:1328–1336.10.1128/EC.00024-06
- Shoji JY, Arioka M, Kitamoto K. Dissecting cellular components of the secretory pathway in filamentous fungi: insights into their application for protein production. Biotechnol. Lett. 2008;30:7–14.
- Higuchi Y, Nakahama T, Shoji JY, Arioka M, Kitamoto K. Visualization of the endocytic pathway in the filamentous fungus Aspergillus oryzae using an EGFP-fused plasma membrane protein. Biochem. Biophys. Res. Commun. 2006;340:784–791.10.1016/j.bbrc.2005.12.077
- Higuchi Y, Arioka M, Kitamoto K. Endocytic recycling at the tip region in the filamentous fungus Aspergillus oryzae. Commun. Integr. Biol. 2009;2:327–328.10.4161/cib.2.4.8385
- Shoji JY, Arioka M, Kitamoto K. Vacuolar membrane dynamics in the filamentous fungus Aspergillus oryzae. Eukaryotic Cell. 2006;5:411–421.10.1128/EC.5.2.411-421.2006
- Russell RC, Yuan HX, Guan KL. Autophagy regulation by nutrient signaling. Cell Res. 2014;24:42–57.10.1038/cr.2013.166
- Fricker MD, Lee JA, Bebber DP, Tlalka M, Hynes J, Darrah PR, Watkinson SC, Boddy L. Imaging complex nutrient dynamics in mycelial networks. J. Microsc. 2008;231:317–331.10.1111/jmi.2008.231.issue-2
- Shoji JY, Arioka M, Kitamoto K. Possible involvement of pleiomorphic vacuolar networks in nutrient recycling in filamentous fungi. Autophagy. 2006;2:226–227.10.4161/auto.2695
- Kikuma T, Arioka M, Kitamoto K. Autophagy during conidiation and conidial germination in filamentous fungi. Autophagy. 2007;3:128–129.10.4161/auto.3560
- Kikuma T, Kitamoto K. Analysis of autophagy in Aspergillus oryzae by disruption of Aoatg13, Aoatg4, and Aoatg15 genes. FEMS Microbiol Lett. 2011;316:61–69.10.1111/fml.2011.316.issue-1
- Yanagisawa S, Kikuma T, Kitamoto K. Functional analysis of Aoatg1 and detection of the Cvt pathway in Aspergillus oryzae. FEMS Microbiol. Lett. 2013;338:168–176.10.1111/fml.2012.338.issue-2
- Shoji JY, Kikuma T, Arioka M, Kitamoto K. Macroautophagy-mediated degradation of whole nuclei in the filamentous fungus Aspergillus oryzae. PLoS ONE. 2010;5:e15650.10.1371/journal.pone.0015650
- Han P, Jin FJ, Maruyama J, Kitamoto K. A large nonconserved region of the tethering protein leashin is involved in regulating the position, movement, and function of Woronin bodies in Aspergillus oryzae. Eukaryotic Cell. 2014;13:866–877.10.1128/EC.00060-14
- Escano CS, Juvvadi PR, Jin FJ, Takahashi T, Koyama Y, Yamashita S, Maruyama J, Kitamoto K. Disruption of the Aopex11-1 gene involved in peroxisome proliferation leads to impaired Woronin body formation in Aspergillus oryzae. Eukaryotic Cell. 2009;8:296–305.10.1128/EC.00197-08
- Bleichrodt RJ, van Veluw GJ, Recter B, Maruyama J, Kitamoto K, Wösten HA. Hyphal heterogeneity in Aspergillus oryzae is the result of dynamic closure of septa by Woronin bodies. Mol. Microbiol. 2012;86:1334–1344.10.1111/mmi.2012.86.issue-6
- Maruyama J, Escaño CS, Kitamoto K. AoSO protein accumulates at the septal pore in response to various stresses in the filamentous fungus Aspergillus oryzae. Biochem. Biophys. Res. Commun. 2010;391:868–873.10.1016/j.bbrc.2009.11.154
- Tanabe Y, Maruyama J, Yamaoka S, Yahagi D, Matsuo I, Tsutsumi N, Kitamoto K. Peroxisomes are involved in biotin biosynthesis in Aspergillus and Arabidopsis. J. Biol. Chem. 2011;286:30455–30461.10.1074/jbc.M111.247338
- Yoon J, Kikuma T, Maruyama J, Kitamoto K. Enhanced production of bovine chymosin by autophagy deficiency in the filamentous fungus Aspergillus oryzae. PLoS ONE. 2013;8:e62512.10.1371/journal.pone.0062512