Abstract
Most cellulases contain carbohydrate-binding modules (CBMs) that largely contribute to their activity for insoluble substrates. Clostridium thermocellum Cel5E is an endoglucanase having xylanolytic activity. The Cel5E originally has a family 11 CBM preferentially binding to β-1,4- and β-1,3-1,4-mixed linkage glucans. In this study, we replaced the CBM with a different type of CBM, either a family 3 microcrystalline cellulose-directed CBM from Clostridium josui scaffoldin, or a family 6 xylan-directed CBM from Clostridium stercorarium xylanase 11A. Chimeric endoglucanases showed enhanced activity that was affected by CBM binding specificity. These chimeric enzymes could efficiently degrade milled lignocellulosic materials, such as corn hulls, because of heterologous components in the plant cell wall, indicating that diverse CBMs play roles in degradation of lignocellulosic materials.
Graphical abstract
Substrate specificities of an endoglucanase, Cel5E from Clostridium thermocellum were affected by fusion of different type CBMs.
Key words:
Introduction
To ensure a reliable future source of energy and raw materials, the utilization of sustainable biomass has considerable advantages over petroleum-based energy sources. Lignocellulosic biomass, obtained as agricultural byproducts and industrial residues, is an abundant, inexpensive, and renewable source of sugars and is a desirable feedstock for the sustainable production of fuels and chemical products. Cellulose and hemicellulose in lignocellulosic biomass can be degraded by microbial enzymes into oligosaccharides or monosaccharides, which are fermentable to biofuels. However, this degradation step is a bottleneck in the process that is preventing establishment of biofuel technology.Citation1) To improve the efficiency of the degradation step, steps to increase the enzymatic activity, for example, by random mutagenesis or site-directed mutagenesis, have been taken. But few researchers have reported examples of mutants showing significantly higher cellulase activity toward insoluble substrates.Citation2)
Lignocellulose degrading enzymes are generally composed of one or more catalytic modules and one or more non-catalytic accessory modules, e.g., a carbohydrate-binding module (CBM). CBMs enhance activity of the enzymes by promoting binding of the catalytic module on the surface of target insoluble substrates.Citation3) Wang et al. reported that artificial Fervidobacterium nodosum Rt17-B1 Cel5As fused with CBMs showed increased cellulase activity for microcrystalline cellulose.Citation4)
Lignocellulose is a recalcitrant material because of its complicated structure containing cellulose, xylan, and other polysaccharides.Citation5) CBMs are highly diverse modules that can be grouped into 69 families on the basis of amino acid sequence similarities and can discriminate the fine structure of polysaccharides in plant cell walls. Thus, it is speculated that replacement of the original CBM in a cellulase with another CBM should switch its specificity for various polysaccharides.
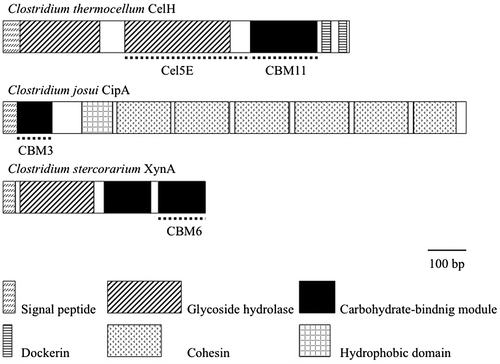
An endoglucanase, Cel5E, from Clostridium thermocellum CelH (CtCel5E), shows endoglucanase and endoxylanase activities. A family 11 CBM (CtCBM11) is appended to the native CtCel5E.Citation6) Here, we show that replacement of CtCBM11 by Clostridium josui CipA CBM 3 or Clostridium stercorarium xylanase 11A CBM 6 changes the specificity of CtCel5E activity.
Materials and methods
Preparation of purified CtCel5E
The genes encoding CtCel5E and CtCel5E-CtCBM11Citation6) were amplified from genomic DNA of C. thermocellum stain 132 and inserted into vector pQE30 (Qiagen, Hilden, Germany), yielding pCtCel5E and pCtCel5E-CtCBM11, respectively. Genes for a family 3 CBM from C. josui, CipA (CjCBM3), and a family 6 CBM from C. stercorarium, xylanase 11A (CsCBM6)Citation7,8) were also amplified and connected downstream of the CtCel5E gene to generate CtCel5E-CjCBM3 and CtCel5E-CsCBM6 sequences (Fig. ). These fusion genes were inserted in pQE30, yielding pCtCel5E-CjCBM3 and pCtCel5E-CsCBM6, respectively. The primers used in this study are shown in supplementary Table 1. These plasmids were expressed in Escherichia coli JM109, and the expressed proteins were purified using a Ni-NTA agarose resin according to the manufacturer’s instructions (Qiagen). Purified proteins were dialyzed against 50 mM potassium phosphate buffer (pH 7.0).
Fig. 1. Isolated cellulase and carbohydrate-binding module genes. Cel5E from C. thermocellum CelH was used as a model enzyme.
Enzymatic activity measurement
CtCel5Es (1.3 μM) was mixed with either 1.5% carboxymethyl cellulose (Sigma-Aldrich, St. Louis, MO, USA), 0.5% ball-milled cellulose, 1% Avicel (Funakoshi, Tokyo, Japan), 0.5% oat-spelt xylan (Fluka, Buchs, Switzerland), or 1% milled corn hulls. The reactions were carried out in 300 μL of 50 mM potassium phosphate buffer (pH 7.0) at 60 °C. The amounts of released reducing sugars were determined by the dinitrosalicylic acid method.Citation9) One unit of activity was defined as moles of glucose equivalents per μmol of enzyme per minute.
Substrate preparations
Carboxymethyl cellulose (Sigma-Aldrich, St. Louis, MO, USA), microcrystalline cellulose (Funakoshi, Tokyo, Japan), and oat-spelt xylan (Fluka, Buchs, Switzerland) were used as commercially available substrates. Ball-milled cellulose was prepared by ball mill processing of 3 g of the KC flock (Nippon Paper Chemicals, Tokyo, Japan) with 1 L of distilled water for 72 h at 4 °C. Corn hull was kindly provided by Dr Khanok Ratanakhanokchai in King Mongkut’s University of Technology Thonburi, Thailand. The corn hull was milled to a final particle size of 1 mm by knife milling.
Results and discussion
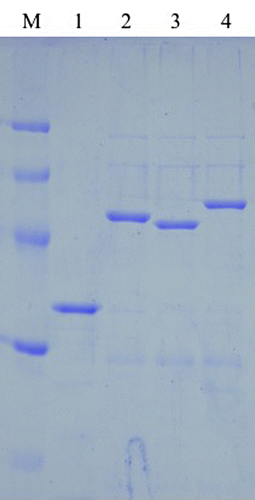
The genes encoding CtCel5E, CtCel5E-CtCBM11, CtCel5E-CjCBM3, and CtCel5E-CsCBM6 were expressed in E. coli JM109, and the resultant proteins were purified with Ni-NTA agarose resin. The purified recombinant protein solutions appeared as a single band following sodium dodecyl sulfate-polyacrylamide gel electrophoresis (Fig. ).
Fig. 2. Purities of recombinant enzymes.
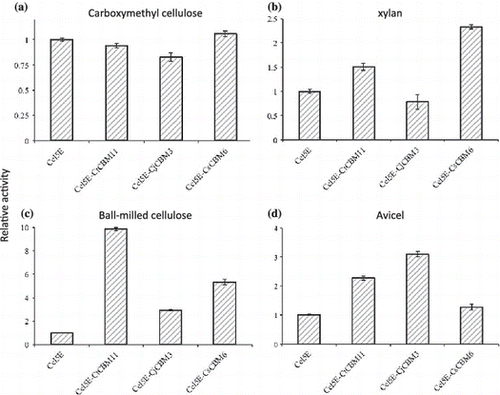
The relative activities of CtCel5Es against substrates are shown in Fig. . There was no significant difference in activity among CtCel5E-CtCBM11, CtCel5E-CsCBM6, and CtCel5E catalytic module for carboxymethyl cellulose, indicating that these CBMs do not affect enzymatic activity for this soluble substrate.Citation3) However, CtCel5E-CjCBM3 showed somewhat lower activity, indicating microcrystalline cellulose-directed CBMs, such as family 1 CBM, affect catalytic activity toward soluble substrates as described by Nozaki et al. Citation10) In contrast, CtCel5E-CtCBM11, CtCel5E-CjCBM3, and CtCel5E-CsCBM6 showed higher activities against insoluble substrates (ball-milled cellulose, Avicel, and oat-spelt xylan, respectively) than the catalytic domain CtCel5E without any CBM (Fig. ). CtCel5E-CtCBM11 showed the highest activity for ball-milled cellulose. CtCel5E-CjCBM3 and CtCel5E-CsCBM6 demonstrated higher activity than CtCel5E-CtCBM11 for Avicel and oat-spelt xylan, respectively, indicating that replacement of the CBM switches the specificity of the catalytic module CtCel5E for insoluble substrates.
Fig. 3. Relative activities of CtCel5E with and without CBMs.
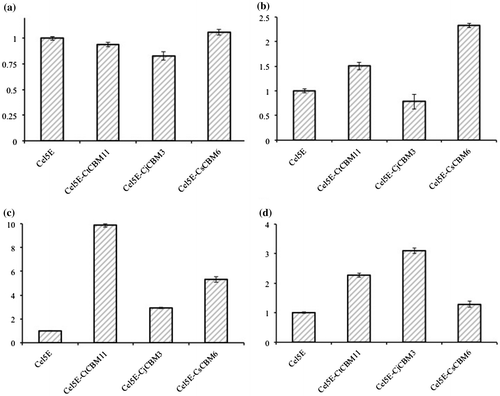
CBMs bind to insoluble polysaccharides and enhance the catalytic activity of their enzymes for insoluble substrates.Citation3) It has been reported that CjCBM3 is not able to bind oat-spelt xylan, whereas CtCBM11 and CsCBM6 do.Citation11–14) The affinity of CtCBM11 and CsCBM6 to oat-spelt xylan is 1.7 × 101 M−1 and 2.3 × 106 M−1, respectively.Citation12–14) This suggests that the quite low affinity of CtCBM11, which should cause low binding of CtCel5E-CtCBM11 to oat-spelt xylan, confers a small increase in CtCel5E activity (Fig. , Suppl. Fig. 1).
The CtCel5E-CtCBM11, CtCel5E-CjCBM3, and CtCel5E-CsCBM6 enzymes showed improvement of CtCel5E activity for ball-milled cellulose and Avicel cellulose (Fig. ). The CtCel5E-CtCBM11 and CtCel5E-CsCBM6 showed higher activities than CtCel5E-CjCBM3 for ball-milled cellulose. It has been reported that CtCBM11,Citation13) CjCBM3,Citation11) and CsCBM6Citation8) bind to insoluble cellulose substrates. Structural studies have reported that each CBM shows a specific binding character due to its unique binding site structure: Family 3 CBM has a flat binding site that preferentially interacts with the surface of crystalline cellulose.Citation14) On the other hand, CtCBM11 has a cleft structure that binds to cellulose, xylan, β-1,3-1,4-glucan, lichenan, and glucomannan.Citation13) CsCBM6 also has a cleft structure that binds to cellulose and xylan.Citation15) These cleft structures can capture a cellulose chain in amorphous regions of cellulose microfibrils. Ball milling of cellulose leads to a considerable reduction in cellulose crystallinity; thus, it converts crystalline cellulose into amorphous cellulose.Citation16) Therefore, it is speculated that larger binding amounts of CtCel5E-CtCBM11 and CtCel5E-CsCBM6 to the ball-milled cellulose lead to the higher activities. Unexpectedly, the binding amounts of CtCel5E-CtCBM11 and CtCel5E-CsCBM6 were lower than that of CtCel5E-CjCBM3 (Suppl. Fig. 1). The binding amounts of these CtCel5Es also did not correlate with their catalytic activity for Avicel cellulose (Suppl. Fig. 1). The structural differences of the CBMs should confer different recognition characteristics for the surface structure of insoluble polysaccharides.Citation17) CtCBM11, CjCBM3, and CsCBM6 might carry CtCel5E to different sites in ball-milled cellulose and Avicel cellulose; thus, fusion enzymes may result in different ratios of improved CtCel5E activity (Fig. ).
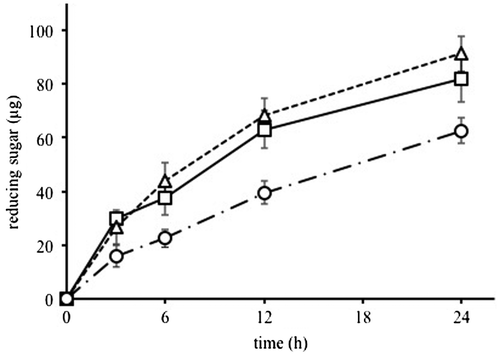
Lignocellulose is a recalcitrant material because of its complicated structure including cellulose, xylan, and other polysaccharides.Citation5) Because each CBM targets different structural sites in plant cell walls, the CBM replacements may enhance the catalytic activity of CtCel5E in complex lignocellulosic materials, as was the case in the improvement of activity toward Avicel cellulose and xylan. Inedible parts of corn are one of the candidates as lignocellulose materials for biofuel.Citation18) The amounts of reducing sugars released from milled corn hulls by the CBM-replaced enzymes, CtCel5E-CjCBM3 and CtCel5E-CsCBM6, were both greater than the amount released by the original enzyme, CtCel5E-CtCBM11 (Fig. ). This result suggests that CBM replacements could enhance catalytic activity of the endoglucanase for lignocellulosic materials including various types of celluloses.Citation19)
Fig. 4. Catalytic activities of CtCel5Es for milled corn hulls.
To improve the efficiency of lignocellulose degradation, attempts have been made to increase the enzymatic activity, but only a few successful results have been reported. We showed that replacement of CtCBM11 with CjCBM3 or CsCBM6 changed the specificity of CtCel5E activity. These CBM-replaced chimeric Cel5Es showed increased activity for Avicel cellulose, xylan and a type of lignocellulosic biomass, milled corn hulls. CBMs having different substrate specificities focus the catalytic module on different sites of lignocellulosic materials and improve their catalytic activity toward polysaccharides in plant cell walls.
Author contribution
S. Karita, M. Kondo, and M. Goto conceived and designed the study; S. Ichikawa and M. Yoshida performed experimental works; S. Ichikawa and S. Karita prepared the manuscript.
Supplementary material
The supplementary material for this paper is available online at http://dx.doi.org/10.1080/09168451.2015.1069696.
Disclosure statement
No potential conflict of interest was reported by the authors.
Funding
This work was supported by a Grant-in-Aid for Scientific Research (C) [grant number 205803062] from the Japan Society for the Promotion of Science.
IchikawaSuppl.Fig1.jpg
Download JPEG Image (240.5 KB)IchikawaSuppl.Table1.jpg
Download JPEG Image (152 KB)References
- Himmel ME, Ding SY, Johnson DK, et al. Biomass recalcitrance: engineering plants and enzymes for biofuels production. Science. 2007;315:804–807.10.1126/science.1137016
- Percival Zhang YH, Himmel ME, Mielenz JR. Outlook for cellulase improvement: screening and selection strategies. Biotechnol. Adv. 2006;24:452–481.10.1016/j.biotechadv.2006.03.003
- Boraston AB, Bolam DN, Gilbert HJ, Davies GJ. Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem. J. 2004;382:769–781.
- Wang Y, Tang R, Tao J, Wang X, Zheng B, Feng Y. Chimeric cellulase matrix for investigating intramolecular synergism between non-hydrolytic disruptive functions of carbohydrate-binding modules and catalytic hydrolysis. J. Biol. Chem. 2012;287:29568–29578.10.1074/jbc.M111.320358
- Burton RA, Gidley MJ, Fincher GB. Heterogeneity in the chemistry, structure and function of plant cell walls. Nat. Chem. Biol. 2010;6:724–732.10.1038/nchembio.439
- Yagüe E, Béguin P, Aubert J-P. Nucleotide sequence and deletion analysis of the cellulase-encoding gene celH of Clostridium thermocellum. Gene. 1990;89:61–67.10.1016/0378-1119(90)90206-7
- Kakiuchi M, Isui A, Suzuki K, et al. Cloning and DNA sequencing of the genes encoding Clostridium josui scaffolding protein CipA and cellulase CelD and identification of their gene products as major components of the cellulosome. J. Bacteriol. 1998;180:4303–4308.
- Sakka K, Kojima Y, Kondo T, Karita S, Ohmiya K, Shimada K. Nucleotide sequence of the Clostridium stercorarium xynA gene encoding xylanase A: identification of catalytic and cellulose binding domains. Biosci. Biotechnol. Biochem. 1993;57:273–277.10.1271/bbb.57.273
- Miller GL. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Biochem. 1959;31:426–428.
- Nozaki K, Nishijima H, Arai T, Mizuno M, Sato N, Amano Y. Regulation of adsorption behavior of carbohydrate-binding module family 1 and endo-β-1,4-glucanase onto crystalline cellulose. J. Appl. Glycosci. 2011;58:133–138.10.5458/jag.jag.JAG-2011_007
- Ichikawa S, Karita S, Kondo M, Goto M. Cellulosomal carbohydrate-binding module from Clostridium josui binds to crystalline and non-crystalline cellulose, and soluble polysaccharides. FEBS Lett. 2014;588:3886–3890.10.1016/j.febslet.2014.08.032
- Bharali S, Purama RK, Majumder A, Fontes CMGA, Goyal A. Functional characterization and mutation analysis of family 11, carbohydrate-binding module (CtCBM11) of cellulosomal bifunctional cellulase from Clostridium thermocellum. Indian J. Microbiol. 2007;47:109–118.10.1007/s12088-007-0023-9
- Carvalho AL, Goyal A, Prates JAM, et al. The family 11 carbohydrate-binding module of Clostridium thermocellum Lic26A-Cel5E accommodates β-1,4- and β-1,3-1,4-mixed linked glucans at a single binding site. J. Biol. Chem. 2004;279:34785–34793.10.1074/jbc.M405867200
- Tormo J, Lamed R, Chirino AJ, et al. Crystal structure of a bacterial family-III cellulose-binding domain: a general mechanism for attachment to cellulose. EMBO J. 1996;15:5739–5751.
- Boraston AB, McLean BW, Chen G, Li A, Warren RAJ, Kilburn DG. Co-operative binding of triplicate carbohydrate-binding modules from a thermophilic xylanase. Mol. Microbiol. 2002;43:187–194.10.1046/j.1365-2958.2002.02730.x
- Yabushita M, Kobayashi H, Hara K, Fukuoka A. Quantitative evaluation of ball-milling effects on the hydrolysis of cellulose catalysed by activated carbon. Catal. Sci. Technol. 2014;4:2312–2317.10.1039/C4CY00175C
- Gilbert HJ. How carbohydrate binding modules overcome ligand complexity. Structure. 2003;11:609–610.10.1016/S0969-2126(03)00103-5
- Youngs H, Somerville C. Plant science. Best practices for biofuels. Science. 2014;344:1095–1096.
- Hromádková Z, Ebringerová A. Isolation and characterization of hemicelluloses of corn hulls. Chem. Papers. 1995;49:97–101.
