Abstract
Expression of Moloney murine leukemia virus (MoMLV)-typed retroviral vectors is strictly suppressed in immature cells such as embryonic stem cells. The phenomenon known as gene silencing is primed by the sequence-specific binding of the zinc finger protein 809 (ZFP809) to the primer-binding site of the vectors. However, it has yet to be determined whether the ZFP809-mediated gene silencing is maintained over a long period. In this study, we established an experimental system that can monitor gene silencing during a long-term cell culture using flow cytometry technology combined with fluorescent reporters for the expression of ZFP809 and the transgene expression driven by the promoters of interest. Time-course analysis using our system revealed that ZFP809 maintains gene silencing effect even at a longtime period. Furthermore, our system was useful for the monitoring of ZFP809-mediated gene silencing regardless of the types of vectors and cell lines.
Graphical abstract
Time-course monitoring of gene silencing
The expression of transgene driven by Moloney murine leukemia (MoMLV) virus is severely suppressed transcriptionally in immature cells such as embryonic stem (ES) cells and hematopoietic stem cells, even though the proviruses are stably integrated in the host genomes.Citation1–3) The phenomenon of attenuation or shutoff of the transgene expression with time, known as gene silencing, is currently considered to be primed by the binding of a nuclear protein to the primer-binding site (PBS) located downstream of the 5′ LTR of the MLV vectors.Citation4–6) In 2009, zinc finger protein (ZFP809), a member of Kruppel-associated box-containing zinc finger proteins (KRAB-ZFPs), was found to be a transcriptional repressor of the MLV vectors.Citation7,8) ZFP809 binds to the repressor-binding site (RBS), an 18-base pair (bp) DNA element overlapping the MLV-derived PBS, resulting in suppression of the transgene expression.Citation8) This was also confirmed in a different context; introduction of a single-nucleotide mutation in the PBS (termed B2 mutation) abolished the binding of ZFP809 to the PBS, and resulted in a resistance of the retroviral vectors to gene silencing in immature cells.Citation9–12)
ZFP809 is known to interact with KAP-1/TRIM28 through its KRAB domain to recruit a H3K9 methyltransferase 4 (ESET, ERG-associated protein with SET domain), heterochromatin protein 1 (HP1), and the NuRD histone deacetylase complex, which leads to a change in the histone modification patterns in the retroviral integration sites.Citation6–8,13) While DNA methylation of retroviral LTRs has been demonstrated to be accompanied by gene silencing,Citation14–16) it was also reported that the gene silencing of retroviral LTR-driven transgene expression was observed in ES cells that are deficient of de novo DNA methyltransferases within two days after transduction.Citation13,17) Since gene silencing has been assessed by the transient expression levels of reporter genes such as luciferases in most of the previous studies,Citation18–21) it was difficult to analyze the extent of it over a longer time period. Retroviral gene silencing is extended over a long term from generation to generation but not transient. Thus, long-term analysis is needed to determine whether the target transcriptional factor has the ability to cause the gene silencing. Furthermore, because our system can monitor the gene silencing by a target transcriptional factor without disrupting cells, our system can analyze the expression and epigenetic modification of the target gene or promoter in time course. In this study, we developed a fluorescent reporter-containing vector system combined with flow cytometry which allows the monitoring of ZFP809-mediated gene silencing events over a long period of time.
Materials and methods
Cell cultures
K562 (a human erythromyeloblastoid leukemia cell line) was purchased from ATCC (American Type Culture Collection) and was cultured in RPMI1640 medium supplemented with 10% fetal calf serum (FCS, Thermo scientific), 2 mM l-glutamine, 0.1 mM sodium pyruvate, 100 U/mL penicillin G sodium, and 100 μg/mL streptomycin sulfate. HEK293 (human embryonic kidney cell line) and 293GP-2 cell lines were purchased from ATCC or TAKARA BIO and were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal calf serum (FCS, Thermo scientific), 2 mM l-glutamine, 0.1 mM sodium pyruvate, 100 U/mL penicillin G sodium, and 100 μg/mL streptomycin sulfate. Ba/F3 (a mouse pro-B-cell line) was cultured in 10% FCS/RPMI added with 10% (v/v) WEHI culture supernatants. F9 (a murine teratocarcinoma stem cell line) cells were obtained from the RIKEN Bio-Resource Center (Tsukuba, Japan) and were cultured in 15% FCS/DMEM. All cell lines were cultured at 37 °C in 5% CO2. 293GPG cells were maintained in DMEM (Sigma-Aldrich) and 10% heat-inactivated FCS (Sigma-Aldrich) supplemented with 0.3 mg/mL G418 (Sigma-Aldrich), 2 μg/mL puromycin (Sigma-Aldrich), and 1 μg/mL tetracycline (Sigma-Aldrich), and cultured at 37 °C in 10% CO2.
Vector construction and preparation
ZFP809 was amplified by reverse transcription (RT)-PCR using the total RNA extracted from F9 cells. These PCR fragments were sub-cloned into pCR2.1 TA cloning vector (The Original TA cloning Kit, Life Technologies). The primer sets used were as follows:
5′-ACGCTCCCAGTCCCATCA-3′ (forward) and
5′-TCAAAAGTACGTTACCCCTGTGTG-3′ (reverse) for ZFP809.
The pCR2.1 TA cloning vector containing truncated ZFP809 was digested with NotI and XhoI, and the fragments were inserted into NotI-XhoI-digested retroviral vector pGCDNsam/IRES/huKO carrying the dl587rev-derived PBS that Matsunari, H. et al. constructed previouslyCitation22) referred to as MSCV/ZFP_huKO. NcoI and ClaI fragments containing EGFP cDNA were cloned into the retroviral vector GCsapMLV carrying the MLV-derived PBSCitation23) referred to as MLV/EGFP.
All vectors were converted to the corresponding retroviruses packaged in VSV-G envelop by transduction into 293GPG cells and then transduced to the target cells with 4 μg/mL of polybrene by spinoculation of 1000 g for 1 h at 32 °C.Citation24)
For the construction of other vectors, fragments of IRES/EGFP cDNA were cloned into the pCMV-SC vector (Agilent) by StrataClone PCR Cloning Kit (Agilent), respectively. The pCMV-SC vector carrying the IRES/EGFP cDNA was further modified by insertion of the MLV- or dl587rev-derived PBS at the downstream of the CMV promoter and referred to as pCMV_MLV/I/EGFP or pCMV_dl587/I/EGFP, respectively. BamHI and HindIII fragments containing the IRES/ EGFP cDNA were cloned into the pBApo-EF1-α Pur DNA vector (TAKARA BIO) and inserted with the MLV- or dl587rev-derived PBS at the downstream of the EF1-α promoter (referred as to pEF1_MLV/I/EGFP or pEF1_dl587/I/EGFP, respectively).
Flow cytometry analysis
Cells were washed and resuspended with phosphate buffered saline containing 2% FSC and analyzed or sorted based on EGFP or huKO expression using FACSAria™ III (BD Biosciences).
RT-PCR
Total RNAs were extracted from cells used FACS analysis (Figs. and ) using the RNeasy Plus Mini Kit (QIAGEN) and converted to cDNAs using SuperScript III First-Strand Synthesis System for RT-PCR (Life Technologies). For amplification of EGFP, RT-PCR was performed using PrimeSTAR GXL DNA Polymerase (TAKARA BIO) and the following oligonucleotides and PCR conditions: 1 min at 98 °C, (10 s at 98 °C, 15 s at 62 °C, and 30 s at 68 °C) × 35 cycle and 5 min at 68 °C. Used primers were as follows.
5′-ATCATGGCCGACAAGCAGAA-3′(forward) and
5′-TCTCGTTGGGGTCTTTGCTC-3′ (reverse) for EGFP.
β-actin was used for a loading control and primers for β-actin were provided with SuperScript III First-Strand Synthesis System.
Bisulfite sequencing
High molecular weight genomic DNAs were obtained from cells sorted by FACS Aria using DNeasy kit (Qiagen) and subjected to the bisulfite PCR using the EpiTect Bisulfite Kit (QIAGEN) and EpiTaq HS (TAKARA BIO). The PCR primers used were as follows:
5′-TGATTTTATGGGATTTTTTTATTG-3′ (forward) and
5′-GAAATTTGGTTTTGTTTTTTTGA-3′ (reverse) for CMV promoter.
5′-GGGGGTAGTTTTAAGTTGGT-3′ (forward) and
5′-CGTCAAAAAAACAAAACCAAATTTCCGA-3′ (reverse) for EF1-α promoter.
5′-AAGGATTTGAAATGATTTTGTGTTT-3′(forward) and
5′-ATCAATCACTCAAAAAAAACCCTC-3′ (reverse) for MLV-LTR.
5′-AAGGATTTGAAATGATTTTGTGTTT-3′ (forward) and
5′-ATCTAAAAAAACCCTCCCAAAAAT-3′ (reverse) for PCMV LTR.
The PCR products were cloned into the TOPO TA Cloning Kit (Life Technologies) and sequenced using the BigDye Terminator V.3.1 Cycle Sequencing Kit and separated on a 3130 Genetic Analyzer (ABI PRISM). Bisulfite sequencing data were analyzed using QUMA (quantification tool for methylation analysis) (http://quma.cdb.riken.jp/).
ChIP-qPCR
ChIP assays were performed using the ChIP Reagents (NIPPON GENE, 318–07131) according to manufacturer’s instructions with modifications. Briefly, cells were fixed with 1% formaldehyde for 5 min. The fixed cells were lysed with SDS lysis buffer (50 mM Tris–HCl pH 8.0, 10 mM EDTA, and 1% SDS) and sonicated using Covaris S220 for chromatin fragmentation (into 150–300-bp fragments). Immunoprecipitation was conducted for 2 h using an antibody specific to H3K9me3 (abcam, ab8898) conjugated to Dynabeads Protein A (Veritas; DB10001), or normal mouse IgG (Millipore, Billerica, MA) conjugated to Dynabeads M-280 Sheep anti-mouse IgG (Veritas; #DB11201) as a negative control. After washing, the beads were incubated in the ChIP direct elution buffer (10 mM Tris–HCl pH 8.0, 300 mM NaCl, 5 mM EDTA, and 0.5% SDS) for 6 h at 65 °C for reverse cross-linking, followed by incubation with 2 μL proteinase K (20 mg/ml) for 2 h at 55 °C. The immune-precipitated DNA was purified using Agencourt AMPure XP beads (Beckman Coulter) according to the manufacturer’s instructions. ChIP assays were performed independently in triplicates. The amount of DNA immunoprecipitated by the antibody was determined by qPCR (LightCycler 480 SYBR Green I Master, Roche Diagnostics) using primer sets:
5′-TACCATGGTGATGCGGTTTT-3′ (forward) and
5′-GGCGGAGTTGTTACGACATT-3′ (reverse) for CMV promoter.
5′-AGAACGGCATCAAGGTGAAC-3′ (forward) and
5′-TGCTCAGGTAGTGGTTGTCG-3′ (reverse) for MLV/EGFP and MSCV/EGFP.
The quantitated values of ChIP DNAs were normalized using the percent input method, in which the value for the ChIP DNA is divided by that of an input sample (1% of starting chromatin) and shown as % input.
Statistical analysis
All statistical analyses were performed by the Student’s t-test.
Results
Establishment of an experimental system for monitoring gene silencing
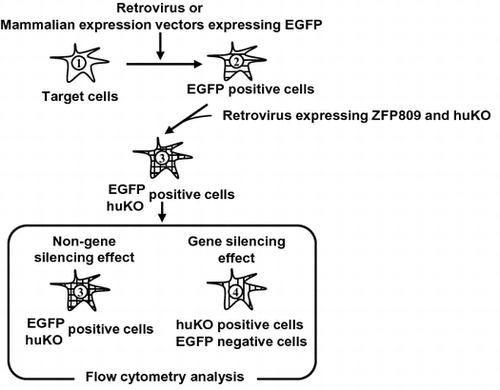
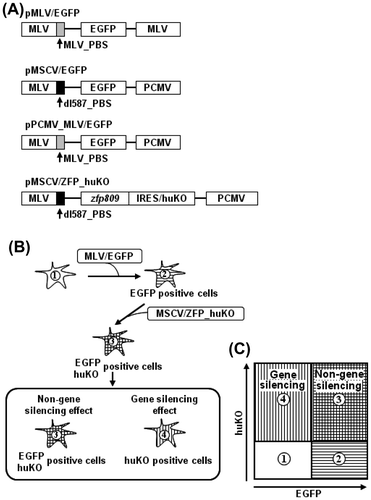
As shown in Fig. (A), we constructed retroviral vectors that carry the MLV- or dl587-derived PBS and the enhanced green fluorescent protein (EGFP) gene (referred to as MLV/EGFP or MSCV/EGFP, respectively) and express ZFP809 and humanized Kusabira Orange (huKO) (referred to as MSCV/ZFP_huKO). The huKO was cloned from the coral stone Fungia concinna and yields an orange-red fluorescence in dimeric form with a 558/583 nm excitation/emission maxima, respectively.Citation25) In addition, an animal transduced with the huKO gene using a retroviral vector stably expressed the huKO gene.Citation22) MSCV-typed vectors have the PBS derived from dl587rev, whose sequence is different at five-bp positions from that of the PBS of MLV and lacks a consensus binding sequence for ZFP809.Citation11) Therefore, we used MSCV-typed vectors as a negative control. Next, target cells were transduced with MLV/EGFP or MSCV/EGFP and sorted on the basis of EGFP expression using FACS Aria (EGFP-positive cells) and then transduced with MSCV/ZFP_huKO (EGFP and huKO double-positive cells) followed by these cells were analyzed by FACS Aria (Fig. (B)). As shown in Fig. (C), FACS analysis was interpreted as below: (1) Negative control of target cells, (2) EGFP single-positive cells were not transduced with MSCV/ZFP_huKO, (3) EGFP and huKO double-positive cells indicated non-gene silencing effect, and (4) huKO single-positive cells were indicated gene silencing effect.
Fig. 1. Schematic diagram of fluorescent reporter-containing vector system combined with flow cytometry methodology.
Time-course monitoring of ZFP809-mediated gene silencing events over a long period
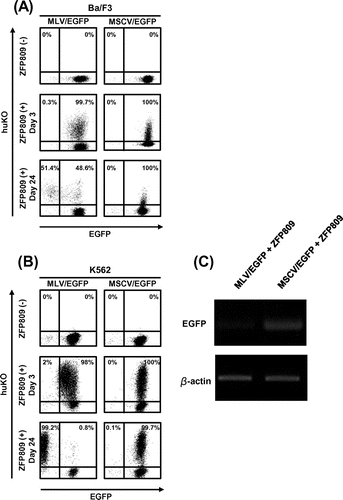
Next, we assessed the effect of ZFP809-mediate gene silencing using our experimental system for a long period of time (for days or weeks) as shown in Fig. (B). We also examined whether our system was useful for suspension cells that have low efficiency of a gene delivery. We used Ba/F3 and K562 cells as suspension cells and transduced these cells with MLV/EGFP or MSCV/EGFP and sorted on the basis of EGFP expression using FACS Aria. These cells were then transduced with MSCV/ZFP_huKO (Fig. ). While EGFP was stably expressed in the cells transduced with MSCV/EGFP even in the presence of ZFP809 expression (the second and third lines from the right in Fig. (A) and (B)), the decline of EGFP expression with time was observed in the cells transduced with MLV/EGFP after infection with MSCV/ZFP_huKO exclusively in the population expressing huKO (the second and third lines from the left in Fig. (A) and (B). Furthermore, we confirmed the repression of GFP expression in cells subjected to FACS analysis by RT-PCR (Fig. (C)).
Fig. 2. Long-term monitoring of ZFP809-mediated gene silencing effect on transgene expression driven by MLV.
Long-term monitoring of ZFP809-mediated gene silencing events on vector carrying CMV and EF1-α promoter with PBS
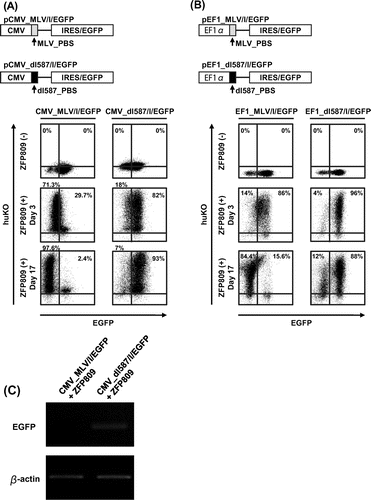
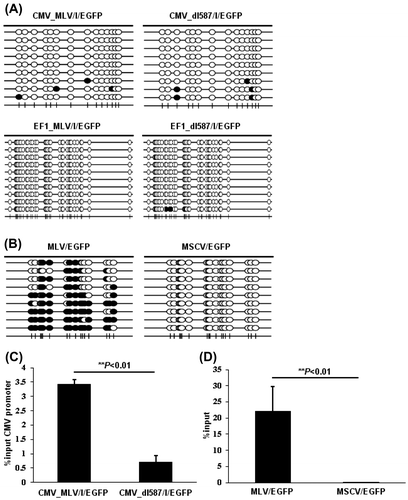
The binding of ZFP809 to the MLV-derived PBS is found to exert transcriptional suppression on non-retroviral promoters containing MLV_PBS.Citation26) We next examined whether events of ZFP809-mediated gene silencing were monitored by our system which used mammalian expression vectors with PBS. We constructed two EGFP expression vectors driven by the CMV promoter containing MLV- or dl587rev-derived PBS (referred to as pCMV_MLV/I/EGFP and pCMV_dl587/I/EGFP, respectively) instead of MLV/EGFP or MSCV/EGFP and transfected each one of them into HEK293 cells together with transduction of MSCV/ZFP_huKO (Fig. (A)). IRES-EGFP was used to enhance GFP expression certainly because the PBS sequence was inserted into downstream of the promoter. As expected, in the HEK293 cells transfected with pCMV_MLV/I/EGFP, EGFP expression declined with time and finally shut off by Day 17, whereas no decline of EGFP expression was observed in the cells transfected with pCMV_dl587/I/EGFP (the second and third lines from the right or left in Fig. (A)). Because the promoter of human elongation factor 1 alpha (EF1-α) gene has been widely used in mammalian expression vectors likewise to the CMV promoter, we also assessed the effect of ZFP809 on the EF1-α promoter (pEF1_MLV/I/EGFP and pEF1_dl587/I/EGFP) by conducting experiments that are parallel with those for the CMV promoter described above (Fig. (B)). Likewise to the CMV promoter, we observed that EGFP expression declined with time in the cells transfected with pEF1_MLV/I/EGFP but not pEF1_dl587/I/EGFP (the second and third lines from the right or left in Fig. (B)). As shown Fig. (C), we also confirmed repression of GFP expression in cells subjected to FACS analysis by RT-PCR.
Fig. 3. Long-term monitoring of ZFP809-mediated gene silencing effect on transgene expression driven by CMV or EF1-α promoters.
Epigenetic modification patterns induced by ZFP809
Since ZFP809 induces repressive histone modifications and de novo DNA methylation at the LTR and the surrounding regions of the vector integration site,Citation13–16,26) we analyzed epigenetic modification in cells subjected to the gene silencing as shown Fig. . We sorted the negative or the positive fraction of EGFP expression in HEK293 cells stably expressing pCMV_MLV/I/GFP or pCMV_dl587/I/GFP and pEF1_MLV/I/GFP or pEF1_dl587/I/GFP together with MSCV/ZFP_huKO using the FACS Aria and assessed the degree of DNA methylation in the promoters by bisulfite PCR followed by clone-based sequencing (Fig. (A)). However, the CpG dinucleotides in the CMV or EF1-α promoter were hardly methylated (Fig. (A)) in contrast to the hyper-methylation observed at the MLV-derived LTR (Fig. (B)) and previous studies.Citation13–16,26) On the other hand, significantly higher levels of H3K9me3 were detected by the ChIP-qPCR analysis at the CMV promoter of pCMV_MLV/I/GFP than that of pCMV_dl587/I/EGFP (Fig. (C)), similar with the H3K9me3 status at the MLV-derived LTR (Fig. (D)) according to previous studies.Citation13–16,26)
Fig. 4. Epigenetic modifications of CMV and EF1-α promoter induced by ZFP809.
Discussion
Gene silencing of transgene driven by retroviral vectors occurs in immature cells despite the successful integration of the transgene into the host chromosomes. Because gene silencing effects are typically assessed by the activities of transiently expressed reporter genes such as luciferase,Citation18–21) it was not possible to determine whether the gene silencing effect was maintained for a long time afterward. Since retroviral gene silencing is extended over a long term, long-term analysis is important for monitoring the gene silencing. To that end, we developed a fluorescent reporter-containing vector system combined with flow cytometry methodology.
Although a gene delivery into suspension cells is typically difficult, our system, which employs the use of retroviral vectors, was useful for these cells and demonstrated that ZFP809-mediated gene silencing was maintained for a long period once it primed the silencing.
Since ZFP809 silences the expression of vectors carrying PBS regardless retroviral vectors,Citation26) we constructed vectors carrying the CMV or EF1-α promoters with PBS (as mammalian expression vectors with PBS) and applied our system to these vectors. Notably, our results demonstrated that the gene silencing was monitored even though the system used mammalian expression vectors and various cell lines. Thus, our system was a useful tool for time course and quantitative assessment of the gene silencing over a longer time period. In addition, our system can analyze the expression and epigenetic modification of target transcriptional factor or promoter because of without disrupting cells. We also confirmed epigenetic modifications such as DNA methylation and histone modification on the CMV and EF1-α promoters. Interestingly, while the ZFP809-mediated silencing of MLV-LTR was accompanied by both K9 trimethylation of Histone H3 and DNA methylation at the LTR,Citation13–16,26) the DNA methylation was not found in the CMV or EF1-α promoters. Nonetheless, the CMV promoter was found to be involved with H3K9 trimethylation. These results suggest that DNA methylation is not essential to ZFP809-mediated gene silencing and varies depending on promoter types. The significance of dual layers of DNA methylation and H3K9 trimethylation in the ZFP809-mediated silencing of MLV-LTR is also highlighted. Retroviruses could transmit vertically from parent to child through germline infection. As a general guard against such transgenerational threats to the host genome, ZFP809 is considered to have evolved to be capable of inducing DNA methylation at MLV-LTR as an epigenetic modification for strong and long-term suppression. Many proteins other than ZFP809 are expected to be involved in gene silencing in mouse and human immature cells. Some of them may regulate the transcription of other types of retroviruses such as HIV-1 and HTLV-1. Recently, two primate-specific KRAB zinc finger genes, ZNF91 and ZNF93, have been reported to repress SVA and L1 retrotransposons in the human genome, respectively.Citation27) Here, we have established a system for monitoring gene silencing events which is applicable for the characterization of other trans-acting factors involved in the silencing of various retroviruses and retrotransposons.
Furthermore, because our system not only can monitor the gene silencing but also investigate the expression trans-acting factors and epigenetic modification at a target promoter, this system is a useful tool for investigating the gene silencing.
Author contributions
Yu Ichida, Yuko Utsunomiya, and Masafumi Onodera conceived and designed the experiments. Yu Ichida and Yuko Utsunomiya performed the experiments. Yu Ichida and Yuko Utsunomiya analyzed the data. Yu Ichida, Yuko Utsunomiya, Junko Tomikawa, Kazuhiko Nakabayashi, Toshinori Sato, and Masafumi Onodera contributed reagents/materials/analysis tools. Yu Ichida, Yuko Utsunomiya, Kazuhiko Nakabayashi, Toshinori Sato, and Masafumi Onodera wrote the paper.
Disclosure statement
No potential conflict of interest was reported by the authors.
Funding
This work was supported by grants from the Ministry of Health, Labor and Welfare and NCCHD.
Acknowledgments
The manuscript was proofread and edited by Dr. Julian Tang of the Department of Education for Clinical Research, NCCHD. We thank all members of department of Human Genetics, NCCHD, and appreciate insightful discussions and comments from Dr. Toru Yasuda, NCCHD.
References
- Akgun E, Ziegler M, Grez M. Determinants of retrovirus gene expression in embryonal carcinoma cells. J. Virol. 1991;65:382–388.
- Cherry SR, Biniszkiewicz D, van Parijs L, Baltimore D, Jaenisch R. Retroviral expression in embryonic stem cells and hematopoietic stem cells. Mol. Cell. Biol. 2000;20:7419–7426.10.1128/MCB.20.20.7419-7426.2000
- Feuer G, Taketo M, Hanecak RC, Fan H. Two blocks in Moloney murine leukemia virus expression in undifferentiated F9 embryonal carcinoma cells as determined by transient expression assays. J. Virol. 1989;63:2317–2324.
- Haas DL, Lutzko C, Logan AC, et al. The Moloney murine leukemia virus repressor binding site represses expression in murine and human hematopoietic stem cells. J. Virol. 2003;77:9439–9450.
- Petersen R, Kempler G, Barklis E. A stem cell-specific silencer in the primer-binding site of a retrovirus. Mol. Cell. Biol. 1991;11:1214–1221.
- Wolf D, Cammas F, Losson R, Goff SP. Primer binding site-dependent restriction of murine leukemia virus requires HP1 binding by TRIM28. J. Virol. 2008;82:4675–4679.10.1128/JVI.02445-07
- Urrutia R. KRAB-containing zinc-finger repressor proteins. Genome Biol. 2003;4:231. doi:10.1186/gb-2003-4-10-231.
- Wolf D, Goff SP. Embryonic stem cells use ZFP809 to silence retroviral DNAs. Nature. 2009;458:1201–1204.10.1038/nature07844
- Barklis E, Mulligan RC, Jaenisch R. Chromosomal position or virus mutation permits retrovirus expression in embryonal carcinoma cells. Cell. 1986;47:391–399. 10.1016/0092-8674(86)90596-9
- Berwin B, Barklis E. Retrovirus-mediated insertion of expressed and non-expressed genes at identical chromosomal locations. Nucleic Acids Res. 1993;21:2399–2407.10.1093/nar/21.10.2399
- Franz T, Hilberg F, Seliger B, Stocking C, Ostertag W. Retroviral mutants efficiently expressed in embryonal carcinoma cells. Proc. Nat. Acad. Sci. USA. 1986;83:3292–3296.10.1073/pnas.83.10.3292
- Loh TP, Sievert LL, Scott RW. Evidence for a stem cell-specific repressor of Moloney murine leukemia virus expression in embryonal carcinoma cells. Mol. Cell. Biol. 1990;10:4045–4057.
- Matsui T, Leung D, Miyashita H, et al. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature. 2010;464:927–931.10.1038/nature08858
- Jähner D, Stuhlmann H, Stewart CL, et al. De novo methylation and expression of retroviral genomes during mouse embryogenesis. Nature. 1982;298:623–628.10.1038/298623a0
- Lorincz MC, Schübeler D, Goeke SC, Walters M, Groudine M, Martin DI. Dynamic analysis of proviral induction and De novo methylation: implications for a histone deacetylase-independent, methylation density-dependent mechanism of transcriptional repression. Mol. Cell. Biol. 2000;20:842–850. 10.1128/MCB.20.3.842-850.2000
- Lorincz MC, Schübeler D, Groudine M. Methylation-mediated proviral silencing is associated with MeCP2 recruitment and localized histone H3 deacetylation. Mol. Cell. Biol. 2001;21:7913–7922.10.1128/MCB.21.23.7913-7922.2001
- Pannell D, Osborne CS, Yao S, et al. Retrovirus vector silencing is De novo methylase independent and marked by a repressive histone code. EMBO J. 2000;19:5884–5894.10.1093/emboj/19.21.5884
- Witzgall R, O’Leary E, Leaf A, Onaldi D, Bonventre JV. The Kruppel-associated box-A (KRAB-A) domain of zinc finger proteins mediates transcriptional repression. Proc. Nat. Acad. Sci. USA. 1994;91:4514–4518.10.1073/pnas.91.10.4514
- Abrink M, Ortiz JA, Mark C, et al. Conserved interaction between distinct Krüppel-associated box domains and the transcriptional intermediary factor 1 beta. Proc. Nat. Acad. Sci. USA. 2001;98:1422–1426.
- Gebelein B. Sequence-specific transcriptional repression by KS1, a multiple-zinc-finger – Krüppel-associated box protein. Society. 2001;21:928–939.
- Mascle XH, Germain-Desprez D, Huynh P, Estephan P, Aubry M. Sumoylation of the transcriptional intermediary factor 1β (TIF1β), the Co-repressor of the KRAB multifinger proteins, is required for its transcriptional activity and is modulated by the KRAB domain. J. Biol. Chem. 2007;282:10190–10202.10.1074/jbc.M611429200
- Matsunari H, Onodera M, Tada N, et al. Transgenic-cloned pigs systemically expressing red fluorescent protein, Kusabira-Orange. Cloning Stem Cells. 2008;10:313–323.10.1089/clo.2008.0024
- Onodera M, Nelson DM, Yachie A, et al. Development of improved adenosine deaminase retroviral vectors. J. Virol. 1998;72:1769–1774.
- Suzuki A, Obi K, Urabe T, et al. Feasibility of ex vivo gene therapy for neurological disorders using the new retroviral vector GCDNsap packaged in the vesicular stomatitis virus G protein. J. Neurochem. 2002;82: 953–960.10.1046/j.1471-4159.2002.01048.x
- Karasawa S, Araki T, Nagai T, Mizuno H, Miyawaki A. Cyan-emitting and orange-emitting fluorescent proteins as a donor/acceptor pair for fluorescence resonance energy transfer. Biochem. J. 2004;381:307–312.
- Rowe HM, Friedli M, Offner S, et al. De novo DNA methylation of endogenous retroviruses is shaped by KRAB-ZFPs/KAP1 and ESET. Development. 2013;140:519–529.10.1242/dev.087585
- Jacobs FM, Greenberg D, Nguyen N, et al. An evolutionary arms race between KRAB zinc-finger genes ZNF91/93 and SVA/L1 retrotransposons. Nature. 2014;516:242–245.10.1038/nature13760
- Negroni M, Buc H. Retroviral recombination: what drives the switch? Nat. Rev. Mol. Cell Biol. 2001;2:151–155.10.1038/35052098
