Abstract
Sake is made from steamed rice, malted rice, and water. Sake production begins with the preparation of a small-scale starter (moto); the quality of moto significantly influences the flavor and richness of sake. In the traditional starter, yamahai-moto, the growth of naturally occurring lactic acid bacteria represses the putrefactive micro-organisms, whereas in the modern starter, sokujo-moto, this is achieved by adding lactic acid. In this study, the successive change in bacterial flora of yamahai-moto was analyzed by pyrosequencing 16S ribosomal RNA genes. Lactobacillus was dominant throughout the process (93–98%). Nitrate-reducing bacteria that have been generally assumed to be the first colonizers of yamahai-moto were scarcely found in the early stage, but Lactobacillus acidipiscis dominated. Lactobacillus sakei drastically increased in the middle stage. This is the first report, though one case study, to show how the early stage microbiota in Japanese yamahai-moto is varyingly controlled without nitrate-reducing bacteria using next-generation sequencing.
Graphical abstract
Microbiota change during the fermentation of traditional Japanese sake starter yamahai-moto.
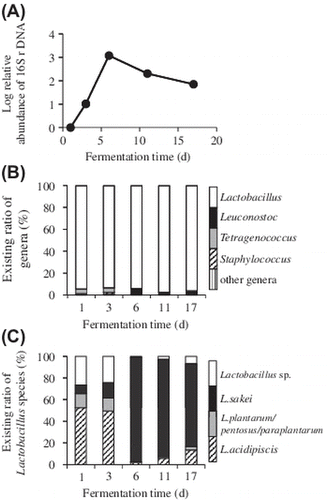
Japanese sake production is conducted by fermenting steamed rice and malted rice (kome-koji) with water.Citation1–4) Kome-koji is a solid-fermented steamed rice prepared using Aspergillys oryzae as a saccharifier to degrade rice starch in the special brewery’s room (koji-muro). Kome-koji is then used for preparing a starter brew, called moto, by manually mixing it with steamed rice and water in an open tank. Modern Japanese sake breweries generally use a type of starter called sokujo-moto, which includes lactic acid to maintain a low pH, thus preventing microbial contamination. Brewer’s yeast Saccharomyces cerevisiae is immediately added, to stimulate alcohol formation.Citation1) Sokujo-moto preparation requires approximately half a month to ensure sufficient viability of yeast to stably ferment the main sake brew (Fig. ). Alternatively, a traditional starter, yamahai-moto (also called kimoto), is made over a longer period of approximately 1 month. In this starter, lactic acid is formed through the growth of lactic acid bacteria (Fig. ). The preparation of yamahai-moto therefore starts at neutral pH, and there is a higher risk of putrefaction by an unexpected growth of spoilage micro-organisms. However, yamahai-moto is still produced by some Japanese breweries, as the sake made using this type of starter has a specific flavor, owing to the complex microbiota succession during the moto preparation.
Fig. 1. Production scheme and generally regarded microbial succession for (A) yamahai-moto and (B) sokujo-moto.
It is known that the early fermentation stage of yamahai-moto contains wild yeasts, film-forming yeasts, and nitrate-reducing bacteria derived from the starting materials and water (Fig. ). Nitrate-reducing bacteria convert nitrate dissolved in the water into nitrite, which suppresses the growth of contaminating micro-organisms. Afterward, lactic acid cocci (Leuconostoc) and bacilli (Lactobacillus) grow, reducing the medium’s pH by lactic acid formation, and suppressing the growth of the above first colonizers. This succession occurs until 10−15 days after the start of moto preparation; at this stage the yeast culture is generally added to produce alcohol. Numerous reports have demonstrated, by culture-dependent microbiota analysis, that Lactobacillus sakei is the dominant species in kimoto and yamahai-moto.Citation2, 5–10) Masuda et al.Citation11) revealed the presence of L. sakei and Leuconostoc mesenteroides in yamahai-moto through denaturing gradient gel electrophoresis of polymerase chain reaction (PCR)-amplified 16S ribosomal RNA gene (rDNA). However, quite recently, Bokulich et al.Citation12) demonstrated, using next-generation 16S rDNA sequencing, that several Lactobacillus species, including Lactobacillus plantarum, were contained in kimoto but that L. sakei was absent in their analyses. To reveal the composition of yamahai-moto microbiota in more detail, culture-independent next-generation sequencing analyses with broad data coverage should be rigorously undertaken.Citation13–17)
We analyzed a sample of yamahai-moto bacterial flora by 16S rDNA-based pyrosequencing. Our data established the dominant bacteria as L. sakei, but, unexpectedly, we could not confirm the presence of nitrate-reducing bacteria, such as Pseudomonas, Achromobacter or Enterobacter, at the early fermentation stage. Based on the results of this one case study and the previous studies, we discuss the possibility of microbial communication in the yamahai-moto fermentation medium.
Materials and methods
Samples
Yamahai-moto was prepared by Shata-shuzo Co. Ltd. (Ishikawa, Japan). Kome-koji and steamed rice were mixed in water taken from a clean well and incubated. The moto was prepared over a four-week period, during which yeast (S. cerevisiae) was added at 13 day. Samples were collected 0–17 days after the start of fermentation for viable counts, acid degree, Baumé degree, pH, and lactic acid concentration; samples taken at 1, 3, 6, 11, and 17 days were used for pyrosequencing analysis. The Baumé degree was measured using a commercially available standard Baumé scale hydrometer, and the acid degree was expressed as the volume (in mL) of 0.1 M sodium hydroxide solution required to neutralize 10 mL of filtered yamahai-moto extract. Lactic acid concentration was measured using a RQflex® plus 10 system with Reflectoquant® lactic acid test strips (Merck).
Pyrosequencing
The variable regions V1 and V2 were used to pyrosequence bacterial 16S rDNA. As reported previously, these regions could be used to classify lactic acid bacteria genera and species.Citation18–22) The primers used for PCR amplification were F8 (5′-AGAGTTTGATCATGGCTCAG-3′, forward) and R357 (5′-CTGCTGCCTYCCGTA-3′, reverse), whose 5′-ends were flanked by specific adaptors (5′-CGTATCGCCTCCCTCGCGCCATCAG-3′ forward, and 5′-CTATGCGCCTTGCCAGCCCGCTCAG-3′ reverse) that are applicable to the pyrosequencing equipment used, namely, the Genome Sequencer FLX® system (Roche, Basel, Switzerland). Short tag sequences (5′-CACGT-3′, 5′-CATAG-3′, 5′-CGATA-3′, 5′-CTATG-3′, and 5′-TACGA-3′) were inserted between the adaptor and primer sequences. These tags allowed reads from different fermentation periods to be sorted in a single run (1, 3, 6, 11, and 17 days after the start of fermentation, respectively). To prepare the template DNA, the samples were suspended in sterilized 0.85% (w/v) sodium chloride solution, followed by vigorous shaking. The resulting suspensions (equivalent to 80 mg yamahai-moto) were used for DNA extraction using EZ-Extract® for DNA (AMR Inc., Gifu, Japan) and a FastPure® DNA kit (Takara Bio, Otsu, Japan). The PCR experiments were performed using PrimeSTAR® GXL DNA polymerase (Takara Bio) with 18–25 cycles of denaturation (94 °C, 30 s), annealing (55 °C, 30 s), and extension (68 °C, 1 min). The average read length (368, 368, 375, 375, and 374 bp for 1, 3, 6, 11, and 17 days, respectively) sufficiently covered the targeted variable regions.
Analysis of pyrosequence reads
Sequence reads were analyzed using the Ribosomal Database Project (RDP) Classifier.Citation23) Reads obtained in the FASTA format were assigned to genus levels with an 80% confidence threshold.Citation24) Chimeric reads were excluded by analyzing the sequences using the DECIPHER server.Citation25,26) Species were determined using the Basic Local Alignment Search Tool (BLAST). Assignment of operational taxonomic units (OTUs), and calculation of the Shannon–Wiener index (taxonomic diversity) were carried out using the FastGroupII© program.Citation27) To determine OTUs, an identity threshold of 97% was set and reads shorter than 300 bp were excluded. Rarefaction curves were drawn using R version 3.1.0 with the vegan package.Citation28,29)
Real-time quantitative PCR
The bacterial population in each sample was determined by real-time quantitative PCR (qPCR). The DNA samples, prepared for pyrosequencing as described, were reused as qPCR templates. The 16S rDNA V3–V4 regions were amplified using 331-f (5′-TCCTACGGGAGGCAGCAGT-3′) and 797-r (5′-GGACTACCAGGGTATCTAATCCTGTT-3′) as forward and reverse primers, respectively.Citation30–32) 5′-FAM-CGTATTACCGCGGCTGCTGGCAC-TAMRA-3′ was used as a TaqMan® probe. Reaction mixtures were prepared using TaqMan® Gene Expression Master Mix (Applied Biosystems, Foster City, CA, USA), and run on a 7300® real-time PCR system (Applied Biosystems), according to the manufacturer’s instructions.
16S rDNA sequence analyses of isolated strains
Bacterial strains were isolated from yamahai-moto at each fermentation stage (0–17 days after the start of fermentation). The strains were cultured on de Man, Rogosa and Sharpe (MRS) agar plates (Becton, Dickinson and Co., Franklin Lakes, NJ, USA), and viable counts were recorded (expressed as colony forming units, cfu, per gram of yamahai-moto). Genomic DNA was extracted from randomly selected isolates using a Wizard® genomic DNA extraction kit (Promega, Fitchburg, WI, USA). Whole regions of 16S rDNA were amplified using the oligonucleotides F8 and R1510 (5′-ACGGYTACCTTTGTTACGACTT-3′) as the primer pair. The amplified fragments were purified using a QIAquick® PCR purification kit (Qiagen, Venlo, Netherlands) and directly sequenced.
Nucleotide sequence accession number
Pyrosequencing reads were deposited in the Sequence Read Archive of the DNA Data Bank of Japan (DDBJ) under accession number DRA002607.
Results
Fermentation during yamahai-moto preparation
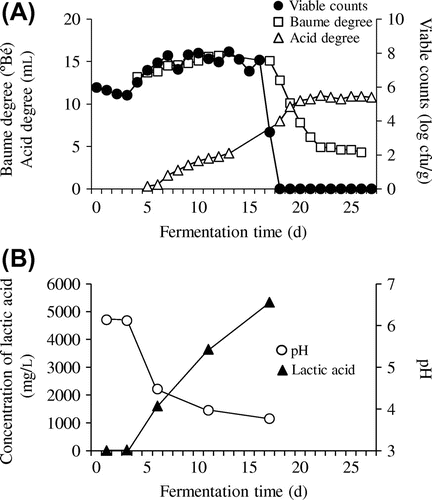
We firstly observed the Baumé and acid degrees and viable counts detected in the time-course samples of yamahai-moto (Fig. (A)). Acid degree significantly increased around 5–8 days after the start of fermentation, with accompanying increase of viable counts on Lactobacillales-selective MRS medium, indicating that lactic acid bacteria contributed to the acid formation. A significant increase in lactic acid concentration (1600 mg/L) and decrease in pH (4.48), observed at 6 day, confirmed this (Fig. (B)). The Baumé degree slightly increased during maturation (until 12 day) but drastically decreased after 17 day, owing to a marked formation of ethanol by the brewer’s yeast added at 13 day (Fig. (A)). Vigorous alcohol formation acted as a bactericide to reduce viable counts to almost zero around 16–18 days; similar succession has been well known in the yamahai-moto fermentation.Citation9) The continuous increase in acid degree after 18 day could therefore be attributed to the action of yeast, not to the previously observed bacteria.
Yamahai-moto microbiota revealed by 16S rDNA pyrosequencing analysis
The samples taken at 1, 3, 6, 11, and 17 day were analyzed for microbiota, as determined by pyrosequencing 16S rDNA. The taxonomic richness and diversity indices are summarized in Table . A marked decrease in species richness (number of OTUs) and diversity (Shannon–Wiener index) were observed at 6 day (OTUs, from 1033 to 668; Shannon–Wiener, from 2.30 to 1.48). These indicated that the drastic taxonomic change took place at this time point.
Table 1. Taxonomic richness and diversity of yamahai-moto samples.
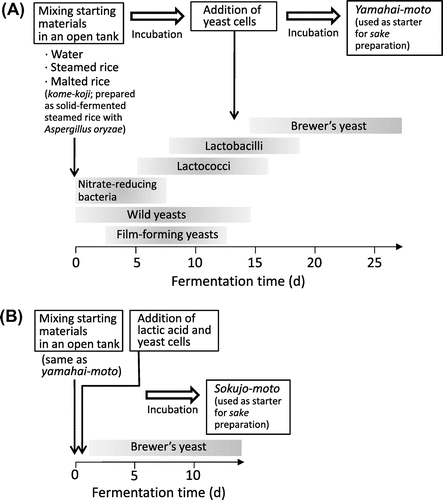
As shown in Table , a broad variety of genera (44 in total) was observed. Rarefaction analysis based on the richness of the genera showed sufficiently saturating curves as shown in Fig. , indicating that enough reads number was sampled from the pyrosequencing data-sets to assess total bacterial community. Throughout the fermentation period, the major constituent was Lactobacillus, which accounted for more than 90% (93–98%, Table and Fig. (B)) of the total population. Species-level BLAST searches revealed that Lactobacillus acidipiscis significantly dominated the microbiota at 1 and 3 days (46–50% of total reads, Fig. (C)), but that the major species afterward was L. sakei (74–91% of total reads, 6, 11, and 17 days, Fig. (C)). A Lactobacillus sp. closely related to L. acidipiscis or pobuzihii formed a secondary significant component at the early stage (23–25%, 1 and 3 days), and Lactobacillus plantarum/pentosus/paraplantarum simultaneously were present in lower proportions (11–12%). Interestingly, a small amount of salt-tolerant lactic acid bacteria Tetragenococcus was detectable in the early stage (4–5%, 1 and 3 days, Fig. (B)), although this yamahai-moto did not include salt. At 6 day, Tetragenococcus decreased and Leuconostoc increased (representing 6% of total population). Staphylococcus was also detected in small amounts in the early stage (2%, 3 day). Notably, nitrate-reducing bacteria known to be included in early stage microbiota (e.g. Pseudomonas, Achromobacter, and Enterobacter, Table ) were scarcely identified (only 1–7 reads of Pseudomonas were detected in each sample).Citation2)
Table 2. Sequence read statistics of yamahai-moto samples and their phylogenetic classification, by RDP classifier.
Fig. 3. Rarefaction curves for each yamahai-moto pyrosequencing data-set.
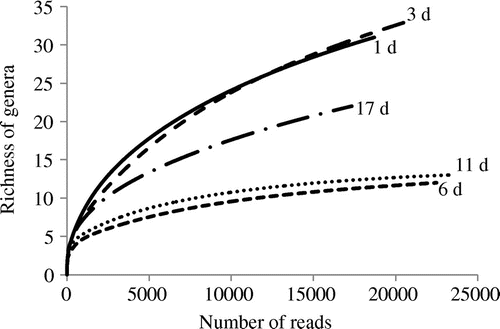
Fig. 4. Microbiota change during the fermentation of yamahai-moto.
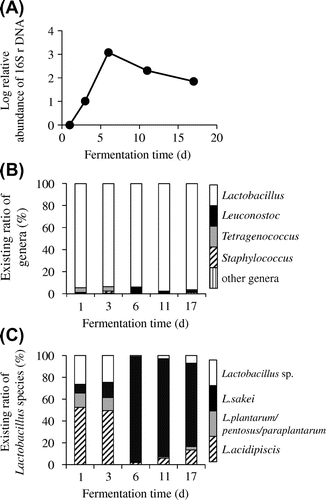
Change of total bacterial population during yamahai-moto fermentation
The qPCR analysis of 16S rDNA indicated a drastic increase in total bacteria 1–6 days from the starting time, corresponding to ~3 on a logarithmic scale (equivalent to a 1000-fold increase, Fig. (A)). This is consistent with the result of viable counts, which increased from approximately 105–106 to 108 cfu/g during the period (Fig. (A)). The population then gradually decreased, but L. sakei remained dominant until 17 day (Fig. (C)). This result indicates that the vigorous growth of L. sakei during the first 6 days is critical in the fermentation of yamahai-moto.
Isolation of bacterial strains from yamahai-moto
We next analyzed the 16S rDNA sequences of bacterial strains isolated on solid MRS medium to identify their species (Table ). Among the 139 strains, the early stage (0–5 days) isolates were predominantly Staphylococcus gallinarum (29 of 43 strains). Other strains were L. sakei (11 strains) and Leuconostoc citreum (3 strains). No Staphylococcus and Leuconostoc strains were isolated at the middle stage (6–11 days); strains grown from the middle and late stage (12–17 days) were solely identified as L. sakei (48 strains for each stage). These results supported those obtained in the pyrosequence survey, which confirmed the rigorous growth of L. sakei until 6 days. All of these members exhibited more than 99% identity with their respective type strains, L. sakei ATCC 15521 (GenBank accession number AM113784), L. citreum ATCC 49370 (AF111948), and S. gallinarum ATCC 35539 (D83366). The sequences of each isolate shared > 99% identity with the pyrosequence reads of the corresponding taxons. Note that we did not identify isolates of L. acidipiscis at any time points. We were unable to discover why this was, but suppose that it may have resulted from culture-dependent approach, which is greatly affected by the growth bias and the limitation of isolate numbers.
Table 3. Taxonomic classification of bacterial strains isolated from yamahai-moto, based on 16S rDNA.
Discussion
This study demonstrated the energetic growth of L. sakei in yamahai-moto fermentation starter. Yamahai-moto fermentation has been regarded as a well-regulated process: (i) the enzymatic actions of A. oryzae contained in kome-koji provide the necessary nutrients, linoleic acid and peptide-forming arginine, to stimulate the growth of L. sakei,Citation33) (ii) nitrate-reducing bacteria form appropriate concentrations of nitrite to repress the growth of wild yeasts, film-forming yeasts, and other putrefactive bacteria in the starting materials.Citation2, 34–36) Along with these events, undesirable micro-organisms gradually decrease their population and Leuconostoc and L. sakei subsequently grow to provide an appropriate acidic condition that is suitable for alcohol fermentation by S. cerevisiae.Citation37) Therefore, initial-stage microbial succession is very important in the yamahai-moto fermentation, as this significantly influences the stability and quality of yamahai-moto.
The microbiota revealed in this study, however, showed a quite different succession; L. acidipiscis was present in the early stage, in the absence of nitrate-reducing bacteria (Table and Fig. ). Actually, we detected little nitrite during yamahai-moto fermentation (less than 0.1 ppm), although considerable amounts have been reported in other studies (up to 27 ppm).Citation34, 38) This indicates that yamahai-moto fermentation has multiple microbiota types, which may be differently controlled with or without nitrite synthesis. It is known that failure in the control of initial microbiota results in a hasty abnormal fermentation, called haya-waki (putrefactive and incomplete maturation produced by wild yeasts)Citation39); perhaps the presence of lactobacilli, possibly L. acidipiscis, in the early stage could prevent putrefaction when nitrites are absent. The 16S rDNA sequence of L. acidipiscis (type strain FS60-1, GenBank accession number AB023836) is largely different from those of the type strains L. sakei ATCC 15521 (AM113784) and L. plantarum ATCC 14917 (D79210) (88% identity with each), therefore it is improbable that the presence of L. acidipiscis in yamahai-moto has been missing throughout the past reports because of the similarity of the 16S ribotype.
Several recent reports demonstrate that the micro-organisms included in the moto fermentation are indigenously regulated, depending on the brewery’s process environment. Bokulich et al.Citation12) suggested, after using a marker gene quantitative PCR approach, that micro-organisms inhabiting the sake brewery’s room surfaces or cellar equipment could essentially influence the fermentation brew microflora. In another report, Masuda et al.Citation11) characterized multiple isolates from a sake fermentation tank surface and moto using pulsed-field gel electrophoresis, and revealed that identical strains of L. mesenteroides were present over several years, and stably grew in that brewery’s moto. Considering these reports, L. acidipiscis and other early stage Lactobacillales detected might have resided in fermentation equipment or on the cellar surface we prepared kome-koji in this study. We isolated a different Leuconostoc species, L. citreum, in this study; this species is known sometimes to replace L. mesenteroides in the moto fermentation.Citation40) It would be interesting to determine whether different Leuconostoc strains inhabit different breweries, thereby establishing the unique flavor and taste of each yamahai-moto.
We previously reported that L. sakei occupied the major part of microbiota in Japanese medieval sushi, kaburazushi, which is made by fermenting salted turnip and salted yellow tail in the presence of kome-koji.Citation22) It is therefore conceivable that L. sakei could originate from kome-koji, which is the only shared ingredient used without undergoing any bactericidal process among yamahai-moto and kaburazushi. Lactic acid bacteria contained in kome-koji could be quite low in number as investigated by Ashizawa and Saito (at not more than 102 /g),Citation41) and major bacteria in kome-koji were Bacillus and Staphylococcus as reported by Bokulich et al.Citation12) Despite the low initial population, the process of moto manufacturing stimulates the vigorous growth of lactic acid bacteria. Future studies should address how the moto lactic acid bacteria are differently controlled to become dominant at the species level, as with the fermentation of other kimotoCitation12) and Korean traditional rice wine takjuCitation42) which contains L. plantarum as its major constituent.
Taken together, this study illustrates that the early yamahai-moto fermentation stage is divided into the nitrite and non-nitrite types. Although the results obtained in this study should carefully be interpreted because they are derived from a case study in one specific brewery’s process environment, the observed microbiota succession indicated that yamahai-moto is more diversely regulated than previously understood.
Author contributions
Conceived and designed the experiments: Takashi Koyanagi, Masashi Kiyohara, Hiroshi Matsui, Takane Katayama, and Hidehiko Kumagai. Performed the experiments: Takashi Koyanagi, Masashi Kiyohara, and Hiroshi Matsui. Analyzed the data: Takashi Koyanagi, Akira Nakagawa, Masashi Kiyohara, and Takane Katayama. Contributed reagents/materials/analysis tools: Atsushi Tsuji, Florin Barla, Harumi Take, Yoko Katsuyama, Koji Tokuda, Shizuo Nakamura, Hiromichi Minami, Toshiki Enomoto. Wrote the paper: Takashi Koyanagi and Takane Katayama.
Disclosure statement
No potential conflict of interest was reported by the authors.
Funding
This work was supported by the City Area Program (Central and Northern Ishikawa Area, 2009) of the Ministry of Education, Culture, Sports, Science, and Technology, Japan.
Acknowledgments
We thank Dr. Jiro Nakayama and Dr. Kenji Sonomoto for their helpful advice on pyrosequencing analysis.
References
- Uemura T, Furusaka C, Sudo T, et al. Studies on the sake yeast. Tohoku J. Agric. Res. 1952;3:185–218.
- Kitagaki H, Kitamoto K. Breeding research on sake yeasts in Japan: history, recent technological advances, and future perspectives. Annu. Rev. Food Sci. Technol. 2013;4:215–235.10.1146/annurev-food-030212-182545
- Kanauchi M. SAKE alcoholic beverage production in Japanese food industry. In: Muzzalupo I, editor. Food industry. Rijeka: InTech; 2013. Chapter 3. Available from: http://www.intechopen.com/books/food-industry/sake-alcoholic-beverage-production-in-japanese-food-industry
- Lee CH, Lee SS. Cereal fermentation by fungi. In: Khachatourians GG, Arora DK, editors. Applied mycology and biotechnology. Vol. 2, Agriculture and food production. Amsterdam: Elsevier; 2002. p. 151–170.
- Iino T, Uchimura T, Komagata K. Characterization of Lactobacillus sakei by the type of stereoisomers of lactic acid produced. J. Gen. Appl. Microbiol. 2003;49:111–121.10.2323/jgam.49.111
- Masuda Y, Takahashi T, Yoshida K, et al. TLR ligands of Lactobacillus sakei LK-117 isolated from seed mash for brewing sake are potent inducers of IL-12. J. Biosci. Bioeng. 2011;112:363–368.10.1016/j.jbiosc.2011.06.007
- Katagiri H, Kitahara K, Fukami K. The characteristics of the lactic acid bacteria isolated from moto, yeast mashes for Saké manufacture. Bull. Agric. Chem. Soc. Japan. 1934;10:156–157.10.1271/bbb1924.10.156
- Yoshizawa K. Sake: production and flavor. Food Rev. Int. 1999;15:83–107.10.1080/87559129909541178
- Ashizawa T. Nihonshu jyozo no shinpi [The mystery of Japanese sake brewing focused on kimoto]. J. Brew. Soc. Japan. 1976;71:424–427. Japanese.
- Ashizawa T, Saito Y. Yamahai shubo ni okeru biseibutsugaku-teki kenkyu (13) [Microbiological research on yamahai-shubo (13)]. J. Brew. Soc. Japan. 1966;61:1033–1036. Japanese.
- Masuda Y, Noguchi T, Takahashi T, et al. DGGE and PFGE analysis of lactic acid bacterial succession during Kimoto making. Seibutsu-Kogaku Kaishi. 2012;90:684–690. Japanese.
- Bokulich NA, Ohta M, Lee M, et al. Indigenous bacteria and fungi drive traditional kimoto Sake fermentations. Appl. Environ. Microbiol. 2014;80:5522–5529.10.1128/AEM.00663-14
- Ercolini D. High-throughput sequencing and metagenomics: moving forward in the culture-independent analysis of food microbial ecology. Appl. Environ. Microbiol. 2013;79:3148–3155.10.1128/AEM.00256-13
- van Hijum SA, Vaughan EE, Vogel RF. Application of state-of-art sequencing technologies to indigenous food fermentations. Curr. Opin. Biotechnol. 2013;24:178–186.10.1016/j.copbio.2012.08.004
- Mayo B, Rachid CT, Alegria A, et al. Impact of next generation sequencing techniques in food microbiology. Curr. Genomics. 2014;15:293–309.10.2174/1389202915666140616233211
- Cocolin L, Alessandria V, Dolci P, et al. Culture independent methods to assess the diversity and dynamics of microbiota during food fermentation. Int. J. Food Microbiol. 2013;167:29–43.10.1016/j.ijfoodmicro.2013.05.008
- Bokulich NA, Mills DA. Next-generation approaches to the microbial ecology of food fermentations. BMB Rep. 2012;45:377–389.10.5483/BMBRep.2012.45.7.148
- Balcázar JL, de Blas I, Ruiz-Zarzuela I, et al. Sequencing of variable regions of the 16S rRNA gene for identification of lactic acid bacteria isolated from the intestinal microbiota of healthy salmonids. Comp. Immunol. Microbiol. Infect. Dis. 2007;30:111–118.10.1016/j.cimid.2006.12.001
- Ninane V, Mukandayambaje R, Berben GJ. Identification of lactic acid bacteria within the consortium of a kefir grain by sequencing 16S rDNA variable regions. AOAC Int. 2007;90:1111–1117.
- Kiyohara M, Koyanagi T, Matsui H, et al. Changes in microbiota population during fermentation of narezushi as revealed by pyrosequencing analysis. Biosci. Biotechnol. Biochem. 2012;76:48–52.10.1271/bbb.110424
- Koyanagi T, Kiyohara M, Matsui H, et al. Pyrosequencing survey of the microbial diversity of ‘narezushi’, an archetype of modern Japanese sushi. Lett. Appl. Microbiol. 2011;53:635–640.10.1111/lam.2011.53.issue-6
- Koyanagi T, Nakagawa A, Kiyohara M, et al. Pyrosequencing analysis of microbiota in Kaburazushi, a traditional medieval sushi in Japan. Biosci. Biotechnol. Biochem. 2013;77:2125–2130.10.1271/bbb.130550
- RDP [Internet]. Michigan: Michigan State University Board of Trustees; c1992–2014. Available from: http://rdp.cme.msu.edu/classifier/classifier.jsp.
- Wang Q, Garrity GM, Tiedje JM, et al. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007;73:5261–5267.10.1128/AEM.00062-07
- Database Enabled Code for Ideal Probe Hybridization Employing R (DECIPHER) [Internet]. Wisconsin: University of Wisconsin-Madison. Available from: http://decipher.cee.wisc.edu/FindChimeras.html.
- Wright ES, Yilmaz LS, Noguera DR. DECIPHER, a search-based approach to chimera identification for 16S rRNA sequences. Appl. Environ. Microbiol. 2012;78:717–725.10.1128/AEM.06516-11
- Yu Y, Breitbar M, McNairnie P, et al. FastGroupII: a web-based bioinformatics platform for analyses of large 16S rDNA libraries. BMC Bioinf. 2006;7:57.10.1186/1471-2105-7-57
- R Foundation for Statistical Computing, R: A language and environment for statistical computing. 2014. Available from: http://www.R-project.org/.
- Oksanen J, Blanchet FG, Kindt R, et al. Vegan: community ecology package. R package version 2.0-10. 2013. Available from: http://CRAN.R-project.org/package=vegan.
- Nadkarni MA, Martin FE, Jacques NA, et al. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology. 2002;148:25–266.
- Santelli CM, Orcutt BN, Banning E, et al. Abundance and diversity of microbial life in ocean crust. Nature. 2008;453:653–656.10.1038/nature06899
- Chakravorty S, Helb D, Burday M, et al. A detailed analysis of 16S ribosomal RNA gene segments for the diagnosis of pathogenic bacteria. J. Microbiol. Methods. 2007;69:330–339.10.1016/j.mimet.2007.02.005
- Yamaji E, Furukawa K, Mizoguchi H, et al. Growth factors required for the predominance of Lactobacillus sakei over Leuconostoc mesenteroides in kimoto. J. Brew. Soc. Japan. 2005;100:281–288. Japanese.10.6013/jbrewsocjapan1988.100.281
- Wakai Y, Shiraishi M, Maenaka M, et al. Isolation and characteristics of nitrate-reducing bacteria in yamahai-moto. Hakkokogaku Kaishi. 1990;68:187–195. Japanese.
- Umezu M, Shibata A, Maeda M. Production of amines by nitrate reducing-bacteria and lactobacilli for sake-brewing. Hakkokogaku kaishi. 1977;55:68–74. Japanese.
- Nishio A, Shigeru K. Control of moto (Sake starter) prepared by traditional method by addition of lactic acid bacteria and nitrate reducing bacteria. Rep. Tottori Inst. Ind. Technol. 2008;11:55–58. Japanese.
- Momose H, Fujikura H. Lactobacilli isolated from moto (Sake starter) prepared by traditional method. J. Brew. Soc. Japan. 1996;91:834–837. Japanese.10.6013/jbrewsocjapan1988.91.834
- Ashizawa T, Saito Y. Yamahai shubo ni okeru biseibutsugaku-teki kenkyu (12) [Microbiological research on yamahai-shubo (12)]. J. Brew. Soc. Japan. 1966;61:638–642. Japanese.
- Mizoguchi H, Hara S. A theoretical consideration of kimoto making. J. Soc. Brew. Japan. 2010;105:124–138. Japanese.10.6013/jbrewsocjapan.105.124
- Kurose N, Asano T, Kawakita S, et al. Isolation and characterization of psychotrophic Leuconostoc citreum isolated from rice koji. Seibutsu-kogaku kaishi. 2004;82:183–190. Japanese.
- Ashizawa T, Saito Y. Yamahai shubo ni okeru biseibutsugaku-teki kenkyu (9) [Microbiological research on yamahai-shubo (9)]. J. Brew. Soc. Japan. 1965;60:803–807. Japanese.
- Kim SY, Yoo KS, Kim JE, et al. Diversity analysis of lactic acid bacteria in Korean rice wines by culture-independent method using PCR-denaturing gradient gel electrophoresis. Food Sci. Biotechnol. 2010;19:749–755.10.1007/s10068-010-0105-z
- Akiyama H. Nihon-Shu [Japanese sake]. Tokyo: Iwanami Shoten; 1994.