Abstract
The transferability of plasmids pCAR1, pB10, R388, and NAH7 was compared using the same donor-recipient system at different cell density combinations in liquid or on a solid surface. pCAR1 was efficiently transferred in liquid, whereas the other plasmids were preferentially transferred on a solid surface. Difference of liquid or solid affected the transfer frequency especially at lower cell densities.
Conjugative transfer of DNA elements, including plasmids, is the most important mechanism of horizontal gene transfer due to its high frequency and ability to disseminate large-sized DNA.Citation 1–3 ) For an in-depth understanding of plasmid transfer in nature, their transfer frequency must be compared under various conditions. Cell density is a key determinant of the behavior of bacterial populations.Citation 4 ) Environmental factors are important factors for bacterial behavior, such as growth on the surface of solid substrate(s) or planktonic cells in liquids or at air–liquid interfaces.Citation 5 ) This study assessed the effect of donor and recipient cell density and mating conditions (liquid or a solid surface) on plasmid transfer frequency.
Bacterial strains were grown overnight in Lysogeny Broth (LB)Citation 6 ) containing appropriate antibiotics at 30 °C. Antibiotics were used at final concentrations of 50 μg mL−1 for kanamycin (Km), 30 μg mL−1 for gentamicin (Gm), 25 μg mL−1 for rifampicin (Rif), 30 μg mL−1 for chloramphenicol (Cm), and 12.5 μg mL−1 for tetracycline (Tc). Solid media were prepared by the addition of 1.6% (w v−1) agar to liquid LB medium. Derivative plasmids of pCAR1,Citation 7–9 ) pB10,Citation 10 ) R388,Citation 11 ) and NAH7Citation 12 ) (pCAR1::rfp,Citation 13 ) pB10::rfp,Citation 14 ) R388::rfp, and NAH7K2,Citation 15 ) respectively) were used in this study. R388::rfp was constructed using a method similar to that described previously for pSM1833,Citation 14,16 ) and Pseudomonas resinovorans CA10dm4RGCitation 17 ) (pCAR1-cured strain, spontaneous Rifr and Gmr gene inserted into chromosome). The rfp-Kmr gene cassette insertion site was determined as described previously,Citation 13 ) and the gene cassette was inserted at position 19965nt of R388 (DDBJ/EMBL/GenBank Accession No. BR000038), an intergenic region between ORF23 (hypothetical protein) and ORF24 (hypothetical protein). The resultant plasmid was transferred from P. resinovorans CA10dm4RG(R388::rfp) into Pseudomonas putida SM1443 (P. putida KT2440-derivative strain)Citation 16 ) and used as a donor of R388::rfp. Similarly, NAH7K2 was transferred from Escherichia coli MVK2Citation 18 ) to P. putida SM1443 and the resultant strain was used as a donor of NAH7K2. P. putida SM1443(pCAR1::rfp)Citation 13 ) and P. putida SM1443(pB10::rfp)Citation 14 ) were used as donors of pCAR1::rfp and pB10::rfp, respectively. P. putida KT2440RGCitation 17 ) (derivative strain of KT2440, spontaneous Rifr and Gmr gene inserted into chromosome) was used as the recipient.
To evaluate the effect of cell density on plasmid transfer frequency, overnight cultures of donor and recipient cells were harvested and washed in fresh LB. The resultant cells were suspended in fresh LB to an optical density at 600 nm (OD600) of 2 × 100, 2 × 10−1, 2 × 10−2, 2 × 10−3, and 2 × 10−4 for the donor, and 2 × 101, 2 × 100, 2 × 10−1, 2 × 10−2, and 2 × 10−3 for the recipient. The ratio of donor to recipient cells was 1:10. For liquid mating, equal volumes (200 μL) of donor and recipient cells were mixed in 2 mL microtubes from which the caps had been removed and the lids sealed with a gas-permeable adhesive seal (Thermo Fisher Scientific). For filter mating (mating on a solid surface), a mixture of donor and recipient cells was transferred onto a 0.22 μm membrane filter (GTBP, 2.5 cm2; Millipore) using Glass Microanalysis Filter Holders and Filtering Flasks (Millipore). The resultant filter was placed on an LB agar plate and incubated at 30 °C for 3 h. After incubation, triplicate aliquots (10 μL) of serially diluted mixtures from each tube were spotted on selective agar plates. Transfer frequencies were calculated by dividing the colony forming units (CFU) mL−1 (liquid) or CFU (cm2)−1 (solid surface) of transconjugants by those of donor cells. All experiments were performed at least twice.
The transfer frequency of plasmids in liquid is shown in Fig. (A). Notably, the transfer frequency of pCAR1::rfp and pB10::rfp between a donor OD600 of 2 × 10−1 and a recipient OD600 of 2 × 100 (#2 in Fig. (A)) was higher than that between a donor OD600 of 2 × 100 and a recipient OD600 of 2 × 101 (#1 in Fig. (A)), despite the identical ratio of donor to recipient cells. The reason why the transfer frequency was low at high-donor density condition was unclear. There might be many donor cells being not paired with recipient cells. Interestingly, the transfer frequencies of R388::rfp and NAH7K2 were below the detection limits at lower cell densities (#5, recipient OD600 = 2 × 10−3 for R388::rfp, #4, recipient OD600 = 2 × 10−2 and #5 for NAH7K2; Fig. (A)). The plasmid transfer frequencies on filters (solid surface) are shown in Fig. (B). pB10::rfp, R388::rfp, and NAH7K2 differed by less than 10-fold according to cell density (Fig. (B)). In comparison, the transfer frequency of pCAR1::rfp differed markedly (more than 100-fold) according to cell density (Fig. (B)). The transfer frequencies of pB10::rfp, R388::rfp, and NAH7K2 on the filters were higher than those in liquid medium irrespective of cell density. By contrast, the transfer frequency of pCAR1::rfp on the filter was lower than that in liquid medium at lower cell densities (#4 and 5, Fig. (A) and (B)). Note that the number of donor cells after mating differed between mating in liquid and on filters (open triangles in Fig. (A) and (B)). The number of donors was ~100-fold larger in liquid at a donor OD600 of 2 × 10−3 or 2 × 10−4 (#4 and #5 in Fig. (A) and (B)). This may have been due to differences in the growth rates of the donor cells between the liquid and solid media. Cell motility on the solid surface was likely restricted in comparison to that in liquid, so cells on the solid surface might experience greater competition for nutrients. Nevertheless, the transfer frequencies of pB10::rfp, R388::rfp, and NAH7K2 were > 10-fold higher on filters than in liquid, while that of pCAR1::rfp was lower on filters (Fig. 1AB). This indicates that the mating conditions (i.e. liquid or solid surface) affected the transfer frequency of pB10, R388, and NAH7, particularly at lower cell densities. This result was probably because donor and recipient cells were more likely to come into contact on the solid surface than in liquid, although this was not the case for pCAR1. At first, we hypothesized that the pili encoded by the plasmids caused this difference. The pilus types of transmissible plasmids are classified into MPFF, MPFG, MPFI, and MPFT based on the proteins used for mating pair formation.Citation 19,20 ) The plasmids pB10, R388 and NAH7 encode MPFT,Citation 19 ) which forms short, rigid pili,Citation 21 ) whereas pCAR1 encodes MPFF as well as F,Citation 19 ) which forms long, flexible pili.Citation 21 ) The transferability of plasmids by long, flexible pili is equivalent in liquids and on solid surfaces, whereas transfer of plasmids by short, rigid pili is more efficient on solid surfaces.Citation 22 ) To assess whether the MPF class influenced plasmid transferability, pDK1KCitation 23 ) and F′tet (Pasteur Institute), both of which have MPFF-type pili, were subjected to mating assays using various cell densities. Although pCAR1, pDK1, and F possess the same MPF class pili, their transfer frequencies in the liquid and on the filters differed markedly (Supplementary Fig. S1). These results indicate that the MPF class might not be a major determinant of plasmid transfer frequency in a liquid or solid medium. The results obtained in this study are partly in contradiction with previous report.Citation 22 ) It is possible that different mating conditions (e.g. donor and recipient strains or different cell densities) caused the different results.
Fig. 1. Transfer frequency of plasmids according to cell density.
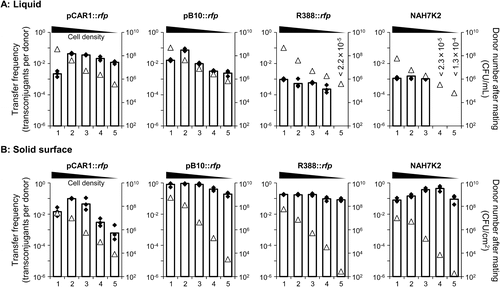
Direct comparisons of the transfer frequency of pCAR1-, pB10-, R388-, and NAH7-derivative plasmids showed that pCAR1 could be efficiently transferred in liquid regardless of donor and recipient cell density. By contrast, R388 and NAH7 showed high transfer frequency on the solid surface, even at lower cell densities. pB10 showed a constant transfer frequency (<100-fold difference) under any conditions. It could be concluded that the mating conditions (liquid or solid medium) affected the transfer frequency of plasmids, especially at lower cell densities. Determination of the effects of various environmental factors on the transferability of plasmids is important to understand not only the influence of the plasmids on the host cell but also the mechanisms by which the plasmids survive in various environments.
Authors contribution
Kosuke Yanagida and Ayako Sakuda carried out the experiments with assistance from Kazuhiro Matsui. Kosuke Yanagida, Chiho Suzuki-Minakuchi, and Masaki Shintani prepared the manuscript. Kazunori Okada contributed analysis and discussion. Hideaki Nojiri designed the project and reviewed the manuscript.
Funding
This work was supported by JSPS KAKENHI [grant number 15H05618] (to M. S.) [grant number 15K14686] (to H. N.).
Supplemental materials
The supplemental material for this paper is available at http://dx.doi.org/10.1080/09168451.2015.1127131.
20151127Supplementary_Figure_S1_final.pdf
Download PDF (147.5 KB)Acknowledgments
We are grateful to Prof. Eva Top of University of Idaho for providing pB10::rfp. We also thank Prof. Masataka Tsuda of Tohoku University for generously providing NAH7K2, pDK1, P. fluorescens Pf-5S, and P. fluorescens Pf-5G.
Disclosure statement
No potential conflict of interest was reported by the authors.
Notes
Abbreviations: Cm, chloramphenicol; Gm, gentamicin; Km, kanamycin; LB, Lysogeny Broth; OD600, optical density at 600 nm; Rif, rifampicin; Tc, tetracycline.
References
- Thomas CM , Nielsen KM . Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 2005;3:711–721.10.1038/nrmicro1234
- Poole AM . Horizontal gene transfer and the earliest stages of the evolution of life. Res. Microbiol. 2009;160:473–480.10.1016/j.resmic.2009.07.009
- Boto L . Horizontal gene transfer in evolution: facts and challenges. Proc. R. Soc. B. 2010;277:819–827.10.1098/rspb.2009.1679
- Toyofuku M , Inaba T , Kiyokawa T , et al . Environmental factors that shape biofilm formation. Biosci. Biotechnol. Biochem. 2015;23:1–6.
- Tashiro Y , Inagaki A , Ono K , et al . Low concentrations of ethanol stimulate biofilm and pellicle formation in Pseudomonas aeruginosa . Biosci. Biotechnol. Biochem. 2014;78:178–181.10.1080/09168451.2014.877828
- Sambrook J , Russell D . Molecular cloning: a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 2001.
- Maeda K , Nojiri H , Shintani M , et al . Complete nucleotide sequence of carbazole/dioxin-degrading plasmid pCAR1 in Pseudomonas resinovorans strain CA10 indicates its mosaicity and the presence of large catabolic transposon Tn4676 . J. Mol. Biol. 2003;326:21–33.10.1016/S0022-2836(02)01400-6
- Takahashi Y , Shintani M , Yamane H , et al . The complete nucleotide sequence of pCAR2: pCAR2 and pCAR1 were structurally identical IncP-7 carbazole degradative plasmids. Biosci. Biotechnol. Biochem. 2009;73:744–746.10.1271/bbb.80665
- Nojiri H . Structural and molecular genetic analyses of the bacterial carbazole degradation system. Biosci. Biotechnol. Biochem. 2012;76:1–18.10.1271/bbb.110620
- Schluter A , Heuer H , Szczepanowski R , et al . The 64 508 bp IncP-1beta antibiotic multiresistance plasmid pB10 isolated from a waste-water treatment plant provides evidence for recombination between members of different branches of the IncP-1beta group. Microbiology. 2003;149:3139–3153.10.1099/mic.0.26570-0
- Fernández-López R , Garcillán-Barcia MP , Revilla C , et al . Dynamics of the IncW genetic backbone imply general trends in conjugative plasmid evolution. FEMS Microbiol. Rev. 2006;30:942–966.10.1111/j.1574-6976.2006.00042.x
- Sota M , Yano H , Ono A , et al . Genomic and functional analysis of the IncP-9 naphthalene-catabolic plasmid NAH7 and its transposon Tn4655 suggests catabolic gene spread by a tyrosine recombinase. J. Bacteriol. 2006;188:4057–4067.10.1128/JB.00185-06
- Shintani M , Fukushima N , Tezuka M , et al . Conjugative transfer of the IncP-7 carbazole degradative plasmid, pCAR1, in river water samples. Biotechnol. Lett. 2008;30:117–122.
- De Gelder L , Vandecasteele FP , Brown CJ , et al . Plasmid donor affects host range of promiscuous IncP-1beta plasmid pB10 in an activated-sludge microbial community. Appl. Environ. Microbiol. 2005;71:5309–5317.10.1128/AEM.71.9.5309-5317.2005
- Ono A , Miyazaki R , Sota M , et al . Isolation and characterization of naphthalene-catabolic genes and plasmids from oil-contaminated soil by using two cultivation-independent approaches. Appl. Microbiol. Biotechnol. 2007;74:501–510.10.1007/s00253-006-0671-4
- Christensen BB , Sternberg C , Andersen JB , et al . Establishment of new genetic traits in a microbial biofilm community. Appl. Environ. Microbiol. 1998;64:2247–2255.
- Shintani M , Habe H , Tsuda M , et al . Recipient range of IncP-7 conjugative plasmid pCAR2 from Pseudomonas putida HS01 is broader than from other Pseudomonas strains. Biotechnol. Lett. 2005;27:1847–1853.10.1007/s10529-005-3892-1
- Miyazaki R , Ohtsubo Y , Nagata Y , et al . Characterization of the traD operon of naphthalene-catabolic plasmid NAH7: a host-range modifier in conjugative transfer. J. Bacteriol. 2008;190:6281–6289.10.1128/JB.00709-08
- Smillie C , Garcillan-Barcia MP , Francia MV , et al . Mobility of plasmids. Microbiol. Mol. Biol. Rev. 2010;74:434–452.10.1128/MMBR.00020-10
- Garcillán-Barcia MP , Alvarado A , de la Cruz F . Identification of bacterial plasmids based on mobility and plasmid population biology. FEMS Microbiol. Rev. 2011;35:936–956.10.1111/j.1574-6976.2011.00291.x
- Lawley TD , Klimke WA , Gubbins MJ , et al . F factor conjugation is a true type IV secretion system. FEMS Microbiol. Lett. 2003;224:1–15.10.1016/S0378-1097(03)00430-0
- Bradley DE , Taylor DE , Cohen DR . Specification of surface mating systems among conjugative drug resistance plasmids in Escherichia coli K-12. J. Bacteriol. 1980;143:1466–1470.
- Yano H , Miyakoshi M , Ohshima K , et al . Complete nucleotide sequence of TOL plasmid pDK1 provides evidence for evolutionary history of IncP-7 catabolic plasmids. J. Bacteriol. 2010;192:4337–4347.10.1128/JB.00359-10
