Abstract
RNA localization is an important event that is essential for the polarization and differentiation of a cell. Although several methods are currently used to detect localized RNAs, a simplified detection system has not yet been developed for Schizosaccharomyces pombe. In the present study, we describe a new vector system for the visualization of localized RNAs in S. pombe using a U1A-tag-GFP system. A pREP1-U1A-tag vector plasmid to express U1A-tagged RNA and a pREP2-U1AGFP plasmid to produce a U1A-GFP fusion protein were constructed for this system. Since the U1A-GFP protein binds U1A-tagged RNA, fluorescence is observed at the location of U1A-tagged RNA in cells expressing both of these. The nucleolar localization of U3 snoRNA was successfully detected using this system, and a novel RNA localized at the DNA region of the nucleus was found by screening localized RNAs. This system will accelerate the study of localized RNAs in S. pombe.
Graphical abstract
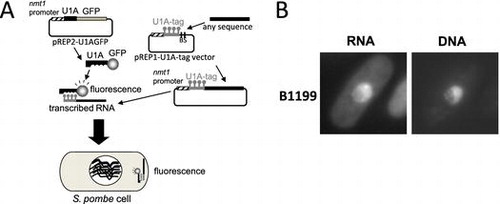
A, A simplified vector system for visualization of localized RNAs in S. pombe. B, A novel RNA localized in the DNA region of the nucleus, B1199, was found by screening.
Key words:
Genetic information written on genomic DNA is copied to RNA by transcription, and is then moved into a cell. Therefore, RNA has the potential to spatially and temporally control the expression of genetic information in a cell. One of the regulatory mechanisms controlling gene expression involves localized RNAs. A large number of RNAs have been reported to localize at specific sites in a cell from bacteria to mammals.Citation1) The localization of mRNA, achieved by active transport, diffusion followed by tethering at specific cellular sites, or local stability, generates cellular polarization and differentiation.Citation1) In Saccharomyces cerevisiae, ASH1 mRNA encoding a transcriptional repressor was shown to be actively transported to the bud-tip of a daughter cell, yielding differences in gene expression between mother and daughter cells.Citation2,3)
Several methods are currently used to detect localized RNAs, and large-scale or genome-wide searches for localized RNAs have been performed in some organisms. In the fluorescence in situ hybridization (FISH) method, probes complementary to RNA sequences are modified to bind a fluorescent protein, hybridized with the RNAs in fixed cells, and detected by fluorescence. In the developing Drosophila embryo, a large-scale FISH analysis was conducted for 3370 genes, and the mRNAs of 71% of these genes showed localization.Citation4) RNA microarray analyses after cell fractionation or the immunoprecipitation of proteins known to bind localized RNAs have also been performed in several organisms. These methods revealed a large number of localized mRNAs, for example, in fractions of the dendrites of rat neurons,Citation5) mitochondriaCitation6) and bud-tips of S. cerevisiae.Citation7) The method using tagged RNA to be bound by the fusion protein of tag-binding proteins and fluorescent proteins has several variations including the MS2 system and U1A system used in living yeast cells.Citation7–10) In this system, a fluorescent-conjugated protein binds to the conformational structure of an RNA tag, such as a hairpin structure. In the U1A system, the RNA recognition motif domain of the U1A protein, associating with U1 snRNA, is fused to the fluorescent protein,Citation11,12) which binds to U1A-tagged RNA to be detected by fluorescence. Large-scale screening of localized RNAs has been performed in S. cerevisiae using the U1A system (U1A-tag-GFP system), and mRNAs localized at the bud-tip or granule in the cytosol were identified in our previous study.Citation13,14)
In addition to localized mRNAs, localized non-coding RNAs have also been investigated. Xist RNAs have been shown to spread on the X chromosome and repress its gene expression.Citation15) NEAT1 is localized at paraspeckles in the nucleus and controls their formation.Citation16) In S. pombe, meiRNA, which is encoded by the sme2+ gene, localizes at the sme2 locus on chromosome II and controls the progression of meiosis.Citation9,17) Although the biological roles of localized non-coding RNAs have been revealed to varying degrees, most have not yet been investigated.
The fission yeast S. pombe is a model organism that has been studied in detail, similar to the budding yeast S. cerevisiae. Both of these yeasts are appropriate for genetic analyses and are good models of higher eukaryotes. However, in the study of RNA, S. pombe is more suitable as a model of higher eukaryotes for the following reasons. While most genes of higher eukaryotes have introns, more than 40% of S. pombe genes, but only 5% of S. cerevisiae genes have introns.Citation18) A homolog of the Dicer protein, which is involved in RNA interferences in higher eukaryotes, is found in S. pombe, but not in S. cerevisiae. Despite the suitability of S. pombe for the study of RNA, localized RNAs have been investigated in less detail in S. pombe than in S. cerevisiae.
In the present study, the U1A-tag-GFP system used to screen localized RNAs in S. cerevisiaeCitation13,14) was applied to S. pombe. This method allowed for the visualization of RNAs in living cells and screening of localized RNAs in S. pombe.
Materials and methods
S pombe strains, media, and genetic methods
The S. pombe strains UR471 (h- leu1-32 ura4-D18) and HM123 (h- leu1-32) were used in the present study. YPD or MM mediumCitation19) with appropriate supplements (uracil, leucine, and thiamine) was used to culture S. pombe cells. The genetic methods used for S. pombe were as previously described.Citation20)
Construction of plasmids and a plasmid library
The oligonucleotides used in this study are listed in Table . pREP2-U1AGFP was made by inserting the Nde I-BamH I PCR fragment amplified with U1A-3 and U1A-4 primers using pT220Citation7) as a template and the 1.0-kb BamH I-Bgl II fragment of pFA6a-GFP(S65T)-kanMX6Citation21) between the Nde I and BamH I-sites of pREP2.Citation22) Prior to the construction of the pREP1-U1A-tag vector, pREP1 was cut by Nde I and Sal I, treated with the klenow fragment, and then self-ligated to yield pREP1A. The Bgl II-BamH I PCR fragment amplified with the U1A-tag-10 and U1A-tag-6 primers using pGAL-U1ACitation7) as a template was ligated to the BamH I site of pREP1A, thereby yielding the pREP1-U1A-tag vector.
Table 1. Oligonucleotides used in the present study.
pREP1-U1A-tag-U3 was constructed by inserting the BamH I-cut PCR fragment amplified with the spU3-3 and spU3-2 primers using S. pombe genomic DNA as a template into the BamH I site of the pREP1-U1A-tag vector. In order to construct the S. pombe plasmid library, S. pombe genomic DNA was partially digested with Sau3A I, and ligated into the BamH I site of the pREP-U1A-tag vector.
pREP1-U1A-tag-B1199-a was constructed by the self-ligation of pREP1-U1A-tag-B1199 cut by Stu I and Sma I. pREP1-U1A-tag-B1199-b was constructed by the self-ligation of pREP1-U1A-tag-B1199 cut with SnaB I and Sma I. pREP1-U1A-tag-B1199-c was constructed by the self-ligation of klenow fragment-treated pREP1-U1A-tag-B1199 cut with Bgl II and Sma I. pREP1-U1A-tag-B1199-d was constructed by the ligation of the 1.1 kb-klenow fragment-treated Eco52 I-Bgl II fragment of pREP1-U1A-tag-B1199 and klenow fragment-treated pREP1-U1A-tag vector cut by BamH I. pREP1-U1A-tag-B1199-e was constructed by the self-ligation of klenow fragment-treated pREP1-U1A-tag-B1199 cut by Eco52 I and Sma I. pREP1-U1A-tag-B1199-r was constructed by the integration of BamH I-cut PCR fragment amplified with B1199-34 and B1199-35 primers using pREP1-U1A-tag-B1199-b as a template, and the inverted sequence was confirmed by the sequence analysis.
In order to make pREP1-LacZ, the LacZ gene of the 3.5-kb Not I fragment of pADβCitation23) was modified at its ends by PCR amplification using LacZ-14 and LacZ-15 primers, and the 0.9-kb Nde I-Cla I, 1.1-kb Cla I-Sac I, and 1.1-kb Sac I-BamH I fragments of the modified LacZ gene were inserted into the Nde I and BamH I sites of pREP1. In order to make pREP1-LacZ-B1199-e, the 1.5-kb Pst I-BamH I fragment of pREP1-B1199-e was exchanged with the 4.3-kb Pst I-BamH I fragment of pREP1-LacZ.
Observation of cells with a fluorescence microscope
UR471 cells carrying the pREP2-U1AGFP and pREP1-U1A-tag vector plasmids with or without an insert sequence were cultured in MM medium supplemented with 25 μg/mL thiamine at 26 °C overnight, washed with MM medium three times to remove thiamine, and incubated at 26 °C for 19 h to the log phase. After Hoechst was added to the culture at a final concentration of 100 μg/mL, cells were harvested and subjected to observations with a fluorescence microscope (Olympus AX70) equipped with a Photometrics Quantix cooled CCD camera.
RT-PCR
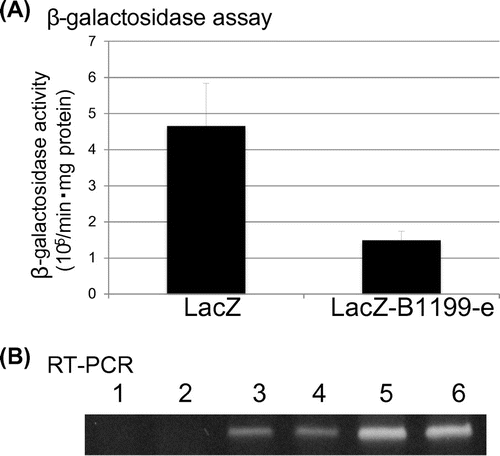
The preparation of RNA and the RT-PCR reaction were performed as described previously.Citation24) U1A-tag-10 and U1A-tag-6 primers were used to amplify the B1199 clones (Fig. (B)). B1199-31 and B1199-32 primers were used in the RT-PCR reaction in Fig. (D). LacZ-3 and LacZ-2 primers were used for the LacZ clones (Fig. (B)). In each assay, the latter primer was used for the RT reaction.
Assay of β-galactosidase
HM123 cells carrying pREP1-LacZ or pREP1-LacZ-B1199-e cultured in MM medium to the log phase were harvested and cell lysates were prepared by disrupting the cells in Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl and 1 mM MgSO4) with glass beads using a vortex mixer. The concentration of total protein in the cell lysates was measured using the Quick Start™ Bradford kit (BIO-RAD). Seven hundred microliters of Z buffer, 1.9 μL β-mercaptoethanol, 100 μL of the cell lysate, and 160 μL of 4 mg/mL ONPG in Z buffer were mixed and incubated at 30 °C. When the color of the mixture turned yellow, 400 μL of 1 M Na2CO3 was added to the mixture and centrifuged at 14,000 rpm for 10 min and the supernatant was subjected to measurements at an absorbance of 420 nm. The activity of β-galactosidase was calculated using the following formula: activity of β-galactosidase = 1,000 × OD420/reaction time (min) × total protein (mg).
Results
Construction of a visualization system for localized RNAs in S. pombe
The U1A-tag-GFP system was tested for its ability to visualize RNAs in S. pombe. In the U1A-tag-GFP system, in which the U1A-GFP fusion protein and U1A-tagged RNA are expressed in a cell, localized RNAs may be observed as fluorescence because the U1A-GFP fusion protein binds the U1A tag (Fig. ). pREP2-U1AGFP and pREP1-U1A-tag vector plasmids were constructed and introduced into S. pombe cells. By removing thiamine from the culture, the U1A-GFP fusion protein and U1A-tag RNA were expressed in the transformed cells. Although slightly stronger fluorescence was observed in the nucleus than in the cytosol, fluorescence was observed throughout the cells by diffusion of the U1A-GFP fusion protein (Fig. ). Cells possessing only pREP2-U1AGFP also showed similar fluorescence patterns (Fig. ).
Fig. 1. The U1A-tag-GFP system in S. pombe.
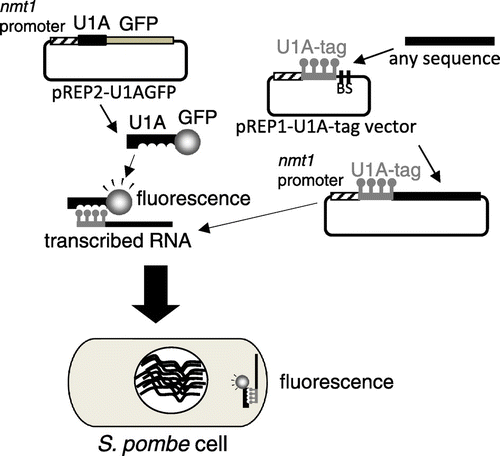
Fig. 2. Visualization of U3 snoRNA localized in the nucleolus and F732 using the U1A-tag-GFP system.
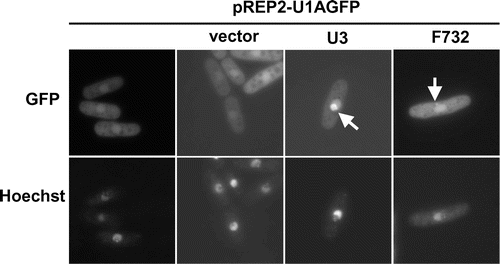
The localization of U3 snoRNA was observed using this system. Cells transformed with pREP2-U1AGFP and pREP1-U1A-tag-U3 showed fluorescence in the nucleolus, located at the non-DNA region in the nucleus (Fig. ). The ratio of cells showing fluorescence in the nucleolar region against the background was approximately 30%, and this was attributed to the diffusion of free U1A-GFP fusion proteins expressed from the strong nmt1 promoter in pREP2-U1AGFP. To reduce the background, pREP42-U1AGFP and pREP82-U1AGFP, which contain the mutated nmt1 promoters with moderate and low transcriptional efficiencies,Citation22) respectively, were constructed; however, the localization of U3 snoRNA was hardly detected using these constructs. pREP1-U1AGFP yielded the best detection results in our study.
Screening of localized RNAs using the U1A-tag-GFP system
A plasmid library was constructed to screen novel localized RNAs. In this library, random fragments of S. pombe genomic DNA were inserted after the U1A-tag sequence of the pREP1-U1A-tag vector. UR471 cells carrying pREP2-U1AGFP were transformed with library plasmids, and we screened for clones showing specific localization in more than 0.5–1% of all cells because the bud-tip localization of ASH1 was observed in 6% of unsynchronized cells in a previous study using the U1A-tag-GFP system in S. cerevisiae.Citation13) In our screening, 6500 transformants with the library plasmid were observed under the fluorescence microscope. While many rRNA clones showed localization in the nucleus, 8 clones showed fluorescence in the DNA region of the nucleus and 1 clone showed localization as a dot in the cytoplasm (Fig. , F732). Of these novel localized RNAs, the B1199 clone, which was isolated first in the screen, was analyzed further.
The B1199 clone showed fluorescence in the DNA region of the nucleus (Fig. (A)), and possessed a 7303-bp fragment of S. pombe genomic DNA (Fig. (B)) after the U1A-tag sequence of the pREP1-U1A-tag vector. In order to identify the localization sequence, shortened versions of the B1199 fragment were made and subjected to observations by the U1A-tag system. As a result, B1199-e possessing a 374-bp fragment showed localization in the DNA region (Fig. (B) and (C)). Interestingly, B1199-r possessing inverted sequence of +1 to +377 nucleotides of B1199 also showed localization in the DNA region (Fig. (B)). The localization of the fluorescence of RNA against the background was observed in more than 10% of all cells.
Fig. 3. Localization of B1199 RNA in the DNA region.
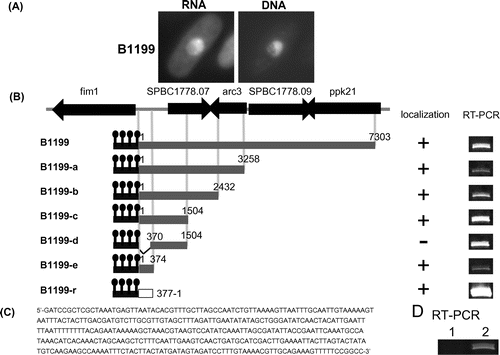
According to the S. pombe genome database, Pombase (www.pombase.org), the 374-base localization sequence of B1199 is located between genes for the transcripts of fim1 and SPBC1778.07 including their 5′-UTRs and 3′-UTRs (Fig. (B)). In order to determine whether the localization sequence of B1199 is expressed in a cell, RT-PCR was performed using the total RNA of wild-type S. pombe cells as a template. As a result, the endogenous expression of the 374-base localization sequence of B1199 was confirmed (Fig. (D)).
Addition of the localization sequence of B1199 to the LacZ gene
pREP1-LacZ-B1199-e, in which the 374-bp fragment of B1199-e was inserted after the stop codon of the LacZ gene, was constructed in order to examine the nuclear localization ability of the 374-bp sequence in B1199-e. If LacZ mRNA with the 374-base sequence of B1199-e is localized in the nucleus, β-galactosidase activity is low because translation to β-galactosidase occurs in the cytoplasm. Cells transformed with pREP1-LacZ or pREP1-LacZ-B1199-e were cultured without thiamine, and the cell lysates extracted were subjected to the β-galactosidase assay. The β-galactosidase activity of cells with pREP1-LacZ-B1199-e was approximately 1/3 that of cells with pREP1-LacZ (Fig. (A)), while the LacZ mRNA levels of these cells were the same (Fig. (B)). This result suggested that mRNA added with the 374-base sequence of B1199-e was more likely to be localized in the nucleus than in the cytoplasm.
Fig. 4. β-galactosidase assay using extracts from cells carrying pREP1-LacZ or pREP1-LacZ-B1199-e.
Discussion
In the present study, the U1A-tag-GFP system designed to visualize localized RNAs successfully detected the nucleolar localization of U3 snoRNA in S. pombe (Fig. ). Furthermore, a novel RNA localized in the DNA region of the nucleus, B1199, was found using this system, indicating its usefulness for the screening of localized RNAs in S. pombe. This is the first study to screen localized RNAs in S. pombe.
However, the biological role of B1199 RNA has not yet been revealed. The 374-base localization sequence of B1199 is located upstream of the reported transcripts of SPBC1778.07 (Fig. (B)). Our preliminary analysis of RT-PCR using primers in B1199-e and the open reading frame of SPBC1778.07 did not yield amplified fragment (data not shown), suggesting low probability that B1199 is a part of the SPBC1778.07 transcript under a general condition. Although the endogenous expression of B1199 RNA was detected by RT-PCR (Fig. (D)), S. pombe cells in which the +44 to +133 sequence of B1199-e in the genome was deleted grew as well as wild-type cells under the several conditions investigated (data not shown). Therefore, further studies are needed to reveal the biological role of B1199 RNA. Eight RNAs localized in the DNA region were identified in our screening. Future investigations on these RNAs will also reveal the localization mechanisms and biological meaning of RNAs localized in the DNA region.
The U1A-tag-GFP system described in the present study is a simplified method for detecting localized RNAs in living S. pombe cells. The localization of an RNA of interest may be easily observed with a fluorescence microscope by cloning the gene of the RNA into the BamH I or Sma I site of the pREP1-U1A-tag vector. Despite the suitability of S. pombe for the study of RNA, investigations of localized RNAs in S. pombe have been insufficient. This simplified system will contribute to accelerating the study of localized RNAs in S. pombe.
Author contributions
Tomoko Takeuchi-Andoh, Sayaka Ohba, and Tokio Tani conceived and designed the experiments. Tomoko Takeuchi-Andoh, Sayaka Ohba, Yu Shinoda, Ayako Fuchita, Sachiko Hayashi, and Emi Nishiyoshi performed the experiments and contributed reagents/materials/analysis tools. Nobuyuki Terouchi made important technical assistance and discussions. Tomoko Takeuchi-Andoh, Sayaka Ohba, and Tokio Tani wrote the paper.
Funding
This work was supported by the Ministry of Education, Culture, Sports, Science and Technology under KAKENHI [grant number 20570005], [grant number 23570011] to TTA.
Acknowledgments
We thank all the members of Tani’s laboratory at Kumamoto University for their helpful discussions.
Disclosure statement
No potential conflict of interest was reported by the authors.
Notes
Abbreviations: ONPG, 2-Nitrophenyl-b-D-galactopyranoside; RT-PCR, reverse transcriptase polymerase chain reaction; bp, base pairs.
References
- Buxbaum AR, Haimovich G, Singer RH. In the right place at the right time: visualizing and understanding mRNA localization. Nat. Rev. Mol. Cell Biol. 2015;16:95–109.
- Long RM, Singer RH, Meng X, et al. Mating type switching in yeast controlled by asymmetric localization of ASH1 mRNA. Science. 1997;277:383–387.10.1126/science.277.5324.383
- Takizawa PA, Sil A, Swedlow JR, et al. Actin-dependent localization of an RNA encoding a cell-fate determinant in yeast. Nature. 1997;389:90–93.
- Lécuyer E, Yoshida H, Parthasarathy N, et al. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell. 2007;131:174–187.10.1016/j.cell.2007.08.003
- Eberwine J, Miyashiro K, Kacharmina JE, et al. Local translation of classes of mRNAs that are targeted to neuronal dendrites. Proc. Natl. Acad. Sci. USA. 2001;98:7080–7085.10.1073/pnas.121146698
- Marc P, Margeot A, Devaux F, et al. Genome-wide analysis of mRNAs targeted to yeast mitochondria. EMBO Rep. 2002;3:159–164.10.1093/embo-reports/kvf025
- Shepard KA, Gerber AP, Jambhekar A, et al. Widespread cytoplasmic mRNA transport in yeast: identification of 22 bud-localized transcripts using DNA microarray analysis. Proc. Natl. Acad. Sci. USA. 2003;100:11429–11434.10.1073/pnas.2033246100
- Beach DL, Salmon ED, Bloom K. Localization and anchoring of mRNA in budding yeast. Curr. Biol. 1999;9:569–578.10.1016/S0960-9822(99)80260-7
- Shichino Y, Yamashita A, Yamamoto M. Meiotic long non-coding meiRNA accumulates as a dot at its genetic locus facilitated by Mmi1 and plays as a decoy to lure Mmi1. Open Biol. 2014;4:140022.10.1098/rsob.140022
- Bertrand E, Chartrand P, Schaefer M, et al. Localization of ASH1 mRNA particles in living yeast. Mol. Cell. 1998;2:437–445.10.1016/S1097-2765(00)80143-4
- Brodsky AS, Silver PA. Pre-mRNA processing factors are required for nuclear export. RNA. 2000;6:1737–1749.10.1017/S1355838200001059
- Takizawa PA, Vale RD, The myosin motor, Myo4p, binds Ash1 mRNA via the adapter protein, She3p. Proc. Natl. Acad. Sci. USA. 2000;97:5273–5278.
- Andoh T, Oshiro Y, Hayashi S, et al. Visual screening for localized RNAs in yeast revealed novel RNAs at the bud-tip. Biochem. Biophys. Res. Commun. 2006;351:999–1004.10.1016/j.bbrc.2006.10.139
- Hayashi S, Andoh T, Tani T. EGD1 (beta-NAC) mRNA is localized in a novel cytoplasmic structure in Saccharomyces cerevisiae. Genes Cells. 2011;16:316–329.10.1111/gtc.2011.16.issue-3
- Briggs SF, Reijo Pera RA. X chromosome inactivation: recent advances and a look forward. Curr. Opin. Genet. Dev. 2014;28:78–82.10.1016/j.gde.2014.09.010
- Clemson CM, Hutchinson JN, Sara SA, et al. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol. Cell. 2009;33:717–726.10.1016/j.molcel.2009.01.026
- Watanabe Y, Yamamoto M. S. pombe mei2+ encodes an RNA-binding protein essential for premeiotic DNA synthesis and meiosis I, which cooperates with a novel RNA species meiRNA. Cell. 1994;78:487–498.10.1016/0092-8674(94)90426-X
- Bähler J, Wood V. The molecular biology of Schizosaccharomyces pombe. Berlin: Springer-Verlag; 2004.
- Gutz H, Heslot H, Leupold U, et al. Handbook of Genetics. New York: Springer Science+Business Media; 1974.
- Alfa C, Fantes P, Hyams J, et al. Experiments with fission yeast. New York, NY: Cold Spring Harbor Laboratory Press; 1993.
- Bähler J, Wu JQ, Longtine MS, et al. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951.10.1002/(ISSN)1097-0061
- Basi G, Schmid E, Maundrell K. TATA box mutations in the Schizosaccharomyces pombe nmt1 promoter affect transcription efficiency but not the transcription start point or thiamine repressibility. Gene. 1993;123:131–136.10.1016/0378-1119(93)90552-E
- MacGregor GR, Caskey CT. Construction of plasmids that express E. coli beta-galactosidase in mammalian cells. Nucleic Acids Res. 1989;17:2365.10.1093/nar/17.6.2365
- Sasaki-Haraguchi N, Ikuyama T, Yoshii S, et al. Cwf16p associating with the nineteen complex ensures ordered exon joining in constitutive pre-mRNA splicing in fission yeast. PLoS One. 2015;10:e0136336.10.1371/journal.pone.0136336
