Abstract
In the course of screening for new anti-influenza virus antibiotics, we isolated herquline A from a culture broth of the fungus, Penicillium herquei FKI-7215. Herquline A inhibited replication of influenza virus A/PR/8/34 strain in a dose-dependent manner without exhibiting cytotoxicity against several human cell lines. It did not inhibit the viral neuraminidase.
Human influenza virus is prevalent worldwide with mutations occurring almost every year. In addition, influenza pandemics occur every few decades on a global scale and can cause millions or 10 millions of deaths.Citation1) Six anti-influenza virus medications with three points of action, the inhibition of neuraminidase, M2 proton channel and RNA polymerase, are currently available. However, problems have already emerged with these medications, including the spread of drug-resistant viruses, differences in receptivity in influenza virus subtypes, and adverse side effects, so novel anti-influenza virus drugs are urgently required. To tackle rapidly evolving and drug-resistant viruses, new medications with significantly different modes of action and chemical structures are actively being sought.Citation2,3)
As a result of our continuous screening for new anti-viral antibiotics from microbial metabolites, we have recently identified new anti-influenza virus diterpene compounds with a novel fused 6/5/6/6 ring skeleton, wickerols A and B, isolated from the culture broth of a fungus, Trichoderma atroviride FKI-3849.Citation4) We have subsequently isolated herquline A from a culture broth of Penicillium herquei FKI-7215, which also possesses potent anti-influenza virus properties in our screening system.Citation4) In this paper, we report the taxonomy of the producing strain, fermentation, isolation, and initial assessment of some bioactive characteristics of herquline A.
The fungal strain FKI-7215 was isolated from a soil sample collected in Niijima, Izu Islands, Tokyo, Japan. The internal transcribed spacer (ITS) sequence of strain FKI-7215 was elucidated and deposited at the DNA Data Bank of Japan with accession number LC043306. The ITS sequence of FKI-7215 was compared to sequences in the GenBank database by BLASTN 2.2.30 analysis.Citation5) The sequence of FKI-7215 was 99.8% similar to that of CBS 336.48 (ex-type of Penicillium herquei; GenBank accession number NR_103659). The producing strain FKI-7215 was thus identified as P. herquei based on its sequence analysis.
One loop of the strain FKI-7215 grown on an LcA slant (0.1% glycerol, 0.08% KH2PO4, 0.02% K2HPO4, 0.02% MgSO4·7H2O, 0.02% KCl, 0.2% NaNO3, and 1.5% agar, pH 6.0) was inoculated into a 500 mL Erlenmeyer flask containing 100 mL of a seed culture medium (2% glucose, 0.2% yeast extract, 0.05% MgSO4·7H2O, 0.5% Polypepton (Wako Pure Chemical Industries, Ltd., Osaka, Japan), 0.1% KH2PO4, and 0.1% agar, pH 6.0) and incubated on a rotary shaker at 27 °C for three days. Ten milliliter of the seed culture was inoculated into each of 10 × 500 mL Erlenmeyer flasks containing a production medium (100 g of water-sodden rice) and cultured statically at 25 °C for six days.
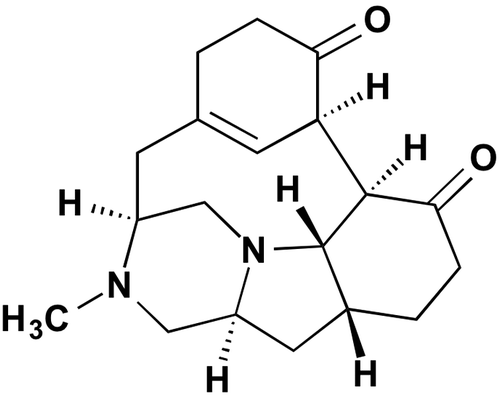
The culture was extracted with 6.0 L of 50% ethanol aq. After filtration, the filtrate was concentrated in vacuo to remove the ethanol. The aqueous solution obtained (3.0 L) was applied to ODS column chromatography and eluted stepwise with 0–100% CH3CN aq. The active fraction (40% CH3CN fraction, 2.0 L) was concentrated in vacuo to remove the CH3CN. The resultant solution (400 mL) was applied to ODS column chromatography and eluted stepwise with 0–100% CH3OH aq. The active fraction (60% CH3OH fraction) was concentrated in vacuo to afford material (220 mg) which was then chromatographed on a silica gel column and eluted stepwise with a mixture of CHCl3-CH3OH (0–100% CH3OH in CHCl3). The active fraction was eluted with a mixture of CHCl3-CH3OH (20:1), collected, and concentrated in vacuo. The concentrated material (39.2 mg) was chromatographed using reverse phase HPLC (Develosil C30-UG, 20 i.d. × 250 mm, Nomura Chemical Co. Ltd., Aichi, Japan) with an isocratic solvent system of CH3CN-H2O (20:80) with 0.1% TFA for 30 min at a flow rate of 7.0 mL/min detected by UV at 220 nm. The peak at the retention time of 19.0 min was collected and concentrated in vacuo to afford 33.4 mg of a compound (1). The structure of 1 was identified as herquline A (Fig. ) by comparison with HPLC analysis and reported NMR and physicochemical property data (Supplemental Table 1 and Fig. 1).Citation6,7) Compound 1 was previously isolated from the same species P. herquei Fg-372.Citation6) Compound 1 has previously been reported by our group as a fungal metabolite via a chemical screening program using Dragendorff’s reagent.Citation6) It was found to inhibit blood platelet aggregation induced by ADP and platelet-activating factor at 180 μM and 240 μM, respectively,Citation7) without acute toxicity (LD50 >100 mg/kg) in mice.Citation6)
The anti-influenza virus activity of 1 was examined by the multi-cycle replication system, in which influenza virus was introduced to influenza virus-susceptible MDCK (Madin-Darby canine kidney) cells, and cell mortality due to the viral infection was evaluated. Cells grown confluently were infected with influenza viruses (A/PR/8/34, A/WSN/33, A/Guizhou/54/89, A/Aichi/2/68 or B/Ibaraki/2/85) at a multiplicity of infection of 0.001 PFU/cell in 200 μL of MEM medium containing 1 mg/mL glucose, 2.25 mg/mL NaHCO3, 3 mg/mL HEPES, 2 mg/mL BSA, 1 mg/L MEM vitamin, 1 mg/L folic acid, 1 mg/L biotin, 0.2 mg/mL gentamicin, 2.5 mg/mL amphotericin B, and 3 μg/mL acetyltrypsin. Test compounds (1, ribavirin, amantadine, zanamivir, or oseltamivir), dissolved in CH3OH at appropriate concentrations, were added and incubated for 72 h at 37 °C under 5% CO2 in a humidified incubator. Anti-influenza virus activity and cell cytotoxicity of the compounds were evaluated by the crystal violet staining method.Citation4) The absorbance (A595) of each well was measured by a microplate reader SH-9000 Lab (Corona Electric Co., Ltd. Ibaraki, Japan.). Ribavirin (viral RNA synthesis inhibitor), amantadine (influenza A virus M2 proton channel blocker), zanamivir (neuraminidase inhibitor), and oseltamivir (neuraminidase inhibitor) were purchased from Sigma-Aldrich (St. Louis, MO, USA).
Compound 1 showed anti-influenza virus activity against the A/PR/8/34 viral strain with an IC50 of 10 μg/mL, without displaying any cytotoxicity in MDCK cells at 500 μg/mL (Table ). Generally, anti-influenza compounds show a broad spectrum of activity against viral strains.Citation8,9) However, 1 demonstrated highly specific anti-influenza virus activity solely against the A/PR/8/34 strain at 10 μg/mL.
Table 1. Anti-influenza virus activity spectrum of 1.
To explore the specificity further, we studied whether 1 influences neuraminidase activity and viral replication. To investigate the effect of 1 on A/PR/8/34 strain neuraminidase activity, we used a fluorescence probe method.Citation9) Neuraminidase activity was measured in 70 μL of PBS (NaCl 8.0 mg/mL, KCl 0.2 mg/mL, Na2HPO4·12H2O 2.9 mg/mL, KH2PO4 0.2 mg/mL), containing 10 μL of 3 μM 4-methylumbelliferyl-N-acetyl-α-D-neuraminic acid (Sigma-Aldrich) and 10 μL of influenza virus (A/PR/8/34, 2.7 × 107 PFU/mL) solution. After addition of 10 μL of 1, zanamivir, or oseltamivir, the reaction mixture was incubated at 37 °C for 10 min. Inhibition (%) of neuraminidase activity was calculated from the fluorescence intensity of cleaved 4-methylumbelliferyl-N-acetyl-α-D-neuraminic acid, which was normalized by control fluorescence intensity of reaction mixture in 70 μL of PBS, containing 10 μL of 1 μM 4-methylumbelliferone (Sigma) and 10 μL of compound solution. The fluorescence intensity was monitored by excitation at 360 nm and emission at 450 nm using a microplate reader SH-9000. We discovered that 1 did not inhibit neuraminidase activity at 300 μg/mL. Positive controls, zanamivir and oseltamivir, inhibited neuraminidase activity at 0.00065 and 11.7 μg/mL, respectively.
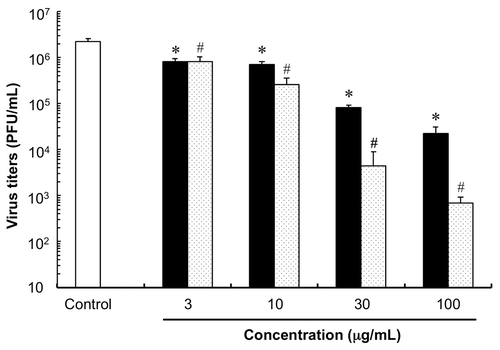
We subsequently tested the effect of 1 on replication of the A/PR/8/34 strain. The plaque assayCitation10,11) was adopted to evaluate the ability of 1 to inhibit the replication of A/PR/8/34 strain. MDCK cells were cultivated in 6-well tissue culture plates with MEM medium at 37 °C under 5% CO2 condition. Confluently growing cells were infected with influenza virus (A/PR/8/34) of serial dilution from 10 to 104 using MEM medium, respectively. MDCK cells were inoculated with influenza virus (A/PR/8/34) at a multiplicity of infection (MOI) of 0.03. After cells were exposed to the virus at room temperature for 30 min, the inoculum was washed with PBS. The cells were overlaid with 1.8 mL of 0.8% nutrient agarose medium (Ina Food Industry Co., Ltd, Nagano, Japan) containing various concentrations of 1 or ribavirin, a known viral RNA synthesis inhibitor, in 4% MeOH solution, and cultured in a humidified incubator for 48 h at 37 °C under 5% CO2 condition. Each well was further overlaid with the agarose medium containing 0.1% neutral red (Sigma-Aldrich) and incubated at 37 °C under 5% CO2 for 3 h. Virus titers were determined by counting the PFU (clear spots) for each sample under an inverted microscope (Olympus Corporation, Tokyo, Japan) and expressed as PFU/mL. As shown in Fig. , the plaque number of A/PR/8/34 was remarkably suppressed by both 1 and ribavirin in a dose-dependent manner. Although, the mechanism of the anti-influenza virus activity of 1 was not elucidated, the molecular target of 1 may be different from that of the other anti-viral drugs.
Fig. 2. Inhibitory activity of 1 against influenza virus replication. Open column = control (4% CH3OH solution); closed column = 1 treatment groups; dotted column = ribavirin treatment groups. Statistical significance of differences between control and compound treatment groups was determined using two groups two-tailed Welch’s t-test. *, #, p < 0.0001 was taken as the level of statistical significance. Each column represents the mean ± SD (n = 6–9).
Finally, to determine the cytotoxic effect of 1, we performed cell growth inhibition assays using several human tumor cell lines. HeLa S3, HT29, A549, H1299, and Panc1 cell lines were cultivated by DMEM medium at 37 °C under 5% CO2 using a humidified incubator. Confluently growing (80%–90%) cells were trypsinized. HeLa S3, HT29, A549, H1299, and Panc1 cell lines were seeded on 96-well microplates (5.0 × 103 cells per well). Test compounds dissolved in MeOH at appropriate concentrations were added to the wells and incubated for 48 h. Resulting live cells were evaluated by the color reaction according to the protocol of WST-8TM (Nacalai Tesque, Inc. Kyoto, Japan). The absorbance (A450) of each well was measured by a Microplate reader SH-9000 Lab. The results showed that 1 did not significantly inhibit cell growth or induce cell death (HeLa S3: IC50 230 μg/mL, HT29: IC50 94 μg/mL, A549: IC50 >300 μg/mL, H1299: IC50 150 μg/mL, and Panc1 cells: >300 μg/mL).
In conclusion, we re-isolated herquline A (1) from the culture broth of Penicillium herquei FKI-7215. We discovered that 1 inhibits influenza virus replication of the A/PR/8/34 strain dose dependently. The molecular target of 1 might be different from that of known anti-influenza virus drugs, and has promise as a new lead compound for antiviral development.
Authors’ contributions
K.S., H.Y., and S.Ō. designed the research plan; T.C., T.N., T.S., Y.W., K.N., and F.M. performed the experiments; T.C., Y.A., and M.I. analyzed the data; and Y.A., K.N., M.I., S.Ō., and K.S. wrote the manuscript.
Supplemental materials
Supplemental material for this article can be accessed at http://dx.doi.org/10.1080/09168451.2016.1162084
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplemental_Section20160222_final.doc
Download MS Word (7.2 MB)Acknowledgments
We are grateful to Dr Kenichiro Nagai and Ms. Noriko Sato, School of Pharmacy, Kitasato University, for help in obtaining NMR and MS data and are grateful to Dr Rokuro Masuma, Kitasato University, for suggestions concerning the experiments.
References
- Neumann G, Noda T, Kawaoka Y. Emergence and pandemic potential of swine-origin H1N1 influenza virus. Nature. 2009;459:931–939.10.1038/nature08157
- Gupta RK, Nguyen-Van-Tam JS. Oseltamivir resistance in influenza A (H5N1) infection. N. Engl. J. Med. 2006;354:1423–1424.
- Jong MD, Bach VC, Phan TQ, et al. Fatal avian influenza A (H5N1) in a child presenting with diarrhea followed by coma. N. Engl. J. Med. 2005;353:2667–2672.10.1056/NEJMoa054512
- Yamamoto T, Izumi N, Ui H, et al. Wickerols A and B: novel anti-influenza virus diterpenes produced by Trichoderma atroviride FKI-3849. Tetrahedron. 2012;68:9267–9271.10.1016/j.tet.2012.08.066
- Altschul SF, Madden TL, Schäffer AA, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402.10.1093/nar/25.17.3389
- Omura S, Hirano A, Iwai Y, Masuma R. Herquline, a new alkaloid produced by Penicillium herquei. Fermentation, isolation and properties. J. Antibiot. 1979;32:786–790.10.7164/antibiotics.32.786
- Enomoto Y, Shiomi K, Hayashi M, et al. Herquline B, a new platelet aggregation inhibitor produced by Penicillium herquei Fg-372. J. Antibiot. 1996;49:50–53.10.7164/antibiotics.49.50
- von Itzstein M. The war against influenza: discovery and development of sialidase inhibitors. Nat. Rev. Drug Discov. 2007;6:967–974.10.1038/nrd2400
- Kim JH, Resende R, Wennekes T, et al. Mechanism-based covalent neuraminidase inhibitors with broad-spectrum influenza antiviral activity. Science. 2013;340:71–75.10.1126/science.1232552
- Hokari R, Nagai T, Yamada H. In vivo anti-influenza virus activity of Japanese herbal (kampo) medicine, "shahakusan," and its possible mode of action. Evid.-based Complement Altern. Med., 2012, 794970.
- Denisova OV, Soderholm S, Virtanen S, et al. Akt inhibitor MK2206 prevents influenza pH1N1 virus infection in vitro. Antimicrob. Agents Chemother. 2014;58:3689–3696.10.1128/AAC.02798-13