Abstract
Carbohydrate isomerases/epimerases are essential in carbohydrate metabolism, and have great potential in industrial carbohydrate conversion. Cellobiose 2-epimerase (CE) reversibly epimerizes the reducing end d-glucose residue of β-(1→4)-linked disaccharides to d-mannose residue. CE shares catalytic machinery with monosaccharide isomerases and epimerases having an (α/α)6-barrel catalytic domain. Two histidine residues act as general acid and base catalysts in the proton abstraction and addition mechanism. β-Mannoside hydrolase and 4-O-β-d-mannosyl-d-glucose phosphorylase (MGP) were found as neighboring genes of CE, meaning that CE is involved in β-mannan metabolism, where it epimerizes β-d-mannopyranosyl-(1→4)-d-mannose to β-d-mannopyranosyl-(1→4)-d-glucose for further phosphorolysis. MGPs form glycoside hydrolase family 130 (GH130) together with other β-mannoside phosphorylases and hydrolases. Structural analysis of GH130 enzymes revealed an unusual catalytic mechanism involving a proton relay and the molecular basis for substrate and reaction specificities. Epilactose, efficiently produced from lactose using CE, has superior physiological functions as a prebiotic oligosaccharide.
Graphical abstract
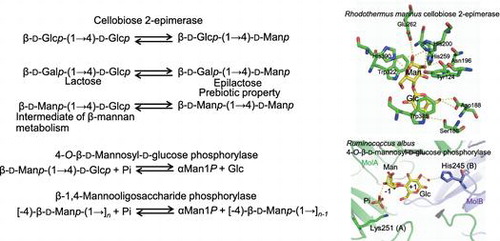
Reactions and substrate recognition of cellobiose 2-epimerase (CE) and mannoside phopshorylases. CE produces prebiotic epilactose. CE epimerizes β-d-Manp-(1→4)-d-Manp for further phosphorolysis.
Carbohydrates are the most abundant chemical compounds in nature. Their structures including monosaccharide components, linkages, chain lengths, and modifications are diverse, and a wide variety of carbohydrate functions are known. They are the major energy source for organisms, and important for forming robust structures in plants, Crustacea, insects, and micro-organisms, resistance to abiotic stresses, and intercellular communication. Several oligosaccharides are known to have beneficial physiological functions, such as prebiotic properties to improve intestinal microfloraCitation1–4) and immune-modulating activity.Citation5,6) A huge number of carbohydrate-metabolizing enzymes, such as glycoside hydrolases, glycosyltransferases, glycoside phosphorylases, and sugar isomerases/epimerases, are involved in the formation and breakdown of carbohydrates. These enzymes are important not only for biological sugar metabolism but also for industrial carbohydrate conversion.
Carbohydrate isomerases and epimerases catalyze the isomerization (interconversion of aldose to ketose, EC 5.3.1.-) and epimerization (interconversion of epimers, EC 5.1.3.-) of carbohydrates, respectively. These enzymes play very important roles in the metabolism of various carbohydrates, including the glycolysis, the oxidative/reductive pentose phosphate pathways, and the Leloir pathway. Among these enzymes, d-xylose isomerase (EC 5.3.1.5), involved in d-xylose metabolism,Citation7,8) is particularly important in the food industry because it is used in the production of d-fructose-rich syrup.Citation9) Furthermore, this enzyme promotes the production of biofuel from lignocellulosic biomass.Citation10) d-Tagatose 3-epimerase (EC 5.1.3.31)Citation11) and d-psicose 3-epimerase (EC 5.1.3.30),Citation12) isolated from Pseudomonas cichorii and Agrobacterium tumefaciens, respectively, made possible the production of a rare sugar, d-psicose, from d-fructose.Citation14) d-Psicose is not used by humans,Citation13) and is expected to be used as a food material with a low glycemic index.Citation15) Furthermore, it exhibits antiobesity activity by increasing energy expenditure.Citation16,17)
Cellobiose 2-epimerase (EC 5.1.3.11; CE), which was first found in a ruminal bacterium Ruminococcus albus, catalyzes the epimerization of a d-glucose residue at the reducing end of cellobiose to a d-mannose residue, i.e. cellobiose is converted to β-d-glucopyranosyl-(1→4)-d-mannose (Glcβ1-4Man).Citation18) CE is the sole enzyme catalyzing the epimerization of oligosaccharides among tens of known carbohydrate isomerases/epimerases. Although CE was found by Tyler and Leatherwood about 50 years ago,Citation18) its structure, function in carbohydrate metabolism, and application to functional oligosaccharides were only reported in the past decade. In this review, recent advances in the study of CE and structurally and functionally related enzymes are addressed.
I. Biochemical functions and distribution of CE
CE from R. albus (RaCE) was successfully purified from cell-free extract of R. albus by column chromatography.Citation19) The molecular mass of RaCE was estimated to be 43 kDa by SDS–PAGE analysis. The enzyme also had epimerization activity toward cellotriose, cellotetraose, lactose, and β-(1→4)-mannobiose (Manβ1-4Man).Citation19–21) β-d-Glucopyranosyl-(1→4)-β-d-glucopyranosyl-(1→4)-d-mannose, β-d-glucopyranosyl-(1→4)-β-d-glucopyranosyl-(1→4)-β-d-glucopyranosyl-(1→4)-d-mannose, epilactose [β-d-galactopyranosyl-(1→4)-d-mannose], and β-d-mannopyranosyl-(1→4)-d-glucose (Manβ1-4Glc) were generated from cellotriose, cellotetraose, lactose, and Manβ1-4Man, respectively. In the reactions with lactose and cellobiose, the conversion levels of epilactose and Glcβ1-4Man were both approximately 30%.Citation22,23) Among β-(1→4)-linked disaccharides, Manβ1-4Man is the best substrate for RaCE in terms of catalytic efficiency (Table ).Citation21) No epimerization activity of RaCE was detectable toward monosaccharides (N-acetyl-d-glucosamine, uridine 5′-diphosphate-d-glucose, d-glucose 6-phosphate, d-glucose, and d-mannose) or glucobioses linked other than by a β-(1→4)-linkage [e.g. maltose: α-(1→4)-linkage; sophorose: β-(1→2)-linkage; laminaribiose: β-(1→3)-linkage; or gentiobiose: β-(1→6)-linkage].Citation20) Thus RaCE is highly specific to a β-(1→4)-linkage at the reducing end of substrates, but can catalyze the conversion of substrates with a d-glucosyl, d-mannosyl, or d-galactosyl residue next to the reducing end sugar residue.
Table 1. Biochemical properties of CEs from various bacterial origins.
Based on the deduced amino acid sequence of RaCE, CE-like genes were found in various genome-sequenced bacteria including anaerobes and aerobes. CEs have been characterized using recombinant enzymes from Bacteroides fragilis,Citation24) Caldicellulosiruptor saccharolyticus,Citation25) Cellulosilyticum lentocellum,Citation26) Cellvibrio vulgaris,Citation27) Dictyoglomus turgidum,Citation28) Dyadobacter fermentans,Citation29) Dysgonomonas gadei,Citation26) Eubacterium cellulosolvens,Citation30) Flavobacterium johnsoniae,Citation29) Herpetosiphon aurantiacus,Citation29) Pedobacter heparinus,Citation29) Rhodothermus marinus,Citation23) Saccharophagus degradans,Citation29) Spirosoma linguale,Citation29) Spirochaeta thermophila,Citation31) and Teredinibacter turneraeCitation29) (Table ). Seventy-one CE-like gene fragments were obtained from environmental DNA (rumen contents, soil, sugar beet extract, and anaerobic sewage sludge) through a PCR-based metagenomics approach, and the biochemical properties of two of them (mD1 and mD2) were investigated (Table ).Citation32) These findings suggest that CE is distributed in various bacteria present in various environments.
All the characterized CEs have optimal pH around neutral, but their optimum temperatures are different depending on the growth temperature of the source organism of the enzyme. CEs from thermophiles such as C. saccharolyticus, D. turgidum, and R. marinus are stable at a high temperature, and show their highest activity at 70–80 °C.Citation23,25,28) Similar to RaCE, these enzymes generally do not use disaccharide substrates other than β-(1→4)-linked disaccharides, although the CEs from D. turgidum and S. thermophila have measurable activity toward maltose and maltotriose.Citation28,31) Kinetic parameters for the epimerization of Manβ1-4Man have been examined with only a few CEs, but the kcat/Km values for this disaccharide are higher than those for cellobiose and lactose.Citation21,27,31,33) This fact is consistent with the prediction that CE is involved in the intracellular metabolism of β-(1→4)-mannooligosaccharides, as described later (see Section III). Cellobiose was used as the substrate when CE was discovered, and this enzyme was named “cellobiose 2-epimerase.” However, considering its substrate selectivity and physiological function, it would be better to call this epimerase “β-(1→4)-mannobiose 2-epimerase” (CE is used throughout this review according to the systematic name of Enzyme Nomenclature). Known CEs generally have higher kcat/Km for cellobiose than for lactose; only R. marinus CE (RmCE) prefers lactose to cellobiose.Citation23) Epilactose, the epimerized product formed from lactose by CE, has beneficial physiological properties as described later (see Section V), and thus RmCE has an attractive substrate selectivity for use as an epilactose-producing enzyme.
Most known CEs catalyze only epimerization reactions; however, surprisingly, the CEs from C. saccharolyticus and D. turgidum also catalyze isomerization (producing epilactose and lactulose from lactose as the epimerization and isomerization products, respectively).Citation25,28) In the reactions of both these enzymes with lactose, conversion levels of epilactose and lactulose finally reached 13–15% and 55–58%, respectively.Citation34) This product distribution is notably different from that of other CEs that catalyze only epimerization.
In spite of high sequence identity with known CEs (32–42%), protein EpiA from C. vulgaris (also called Cellvibrio mixtus) has been considered to be an epimerase converting d-mannose to d-glucose.Citation35) However, biochemical analysis clearly demonstrated that this enzyme has a much higher preference for disaccharide substrates including Manβ1-4Man than d-mannose (kcat/Km for Manβ1-4Man is 5.5 × 104-fold higher than for d-mannose), indicating that C. vulgaris EpiA is a typical CE.Citation27) It is worth mentioning that the activity toward β-(1→4)-mannotriose is 0.24% of that toward Manβ1-4Man. This substrate selectivity is highly consistent with that of the CEs from D. turgidum and S. thermophila.Citation28,31) These findings suggest that CE has high disaccharide selectivity, although epimerization activity toward oligosaccharides longer than a disaccharide was reported for several CEs.Citation19,24,29) Interestingly, C. saccharolyticus CE has low substrate selectivity. Its activities toward monosaccharide substrates, d-glucose, d-mannose, d-xylose, d-lyxose, and d-fructose, are 4.2, 1.1, 1.7, 1.0, and 0.26% of that toward cellobiose, respectively.Citation25)
II. Structure–function relationship of CE
Amein and Leatherwood reported that Glcβ1-4Man, produced from cellobiose by CE in D2O, has deuterium at the C2 position of the reducing end d-mannose residue,Citation36) indicating that the CE reaction proceeds via a cis-enediol intermediate generated by the abstraction of 2H from the reducing end sugar moiety. Structural analysis of RaCE and RmCE revealed that both these CEs have an (α/α)6-barrel-fold catalytic domain (Fig. (A)).Citation37,38) The catalytic domain and active site of CEs are similar to those of N-acylglucosamine 2-epimerases (EC 5.1.3.8; AGE)Citation39,40) and aldose-ketose isomerases (AKI; catalyzing interconversion of d-glucose, d-mannose, and d-fructose)Citation41) with root mean square deviation values of 3.2 and 2.7 Å, respectively, although the overall amino acid sequence similarity between the CEs and these enzymes is low. Substrate and reaction specificities of CEs, AGEs, and AKIs are very different, but their structural similarity indicates that they share common catalytic machinery and mechanism, especially in the formation of the cis-enediol intermediate.
Fig. 1. Structure–function relationship of CE.
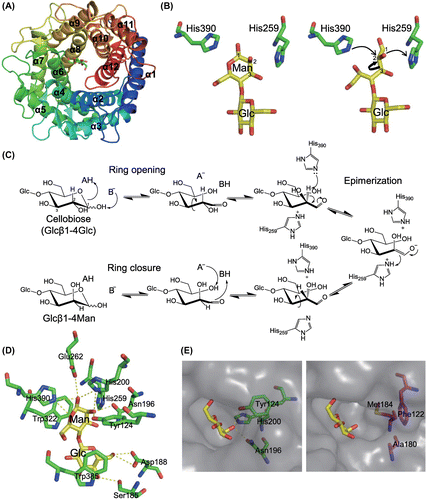
Based on structural and mutational analysis of Anabaena sp. CH1 AGE, Lee et al. proposed a proton abstraction and donation mechanism involving two catalytic His residues (His239 and His372).Citation40) These His residues correspond to the His residues of pig kidney AGE (His248 and His382), predicted from the crystal structure to be essential for catalysis because they are close to an undefined substrate ligand.Citation39) His243 and His374 of RaCE, corresponding to these two essential His residues of AGEs, were shown to be essential for catalytic activity by site-directed mutational analysis.Citation42) The structure of a complex of RmCE and cellobiitol, which is an open-form substrate analog, resulted in a better understanding of the functions of the two catalytic His residues in the proton abstraction/donation mechanism (Fig. (B)).Citation38) The d-glucitol part of cellobiitol bound to the enzyme in a cis-enediol-like conformation in which the O2 atom of the d-glucitol part was rotated around the C2–C3 bond by about 90° from the equatorial position of the O2 atom in the ideal conformation of cellobiose. The C1–C2 bond of the d-glucitol part of cellobiitol was positioned between the two catalytic His residues (His259 and His390). This structure was considered to be the state just before the abstraction of the H2 proton. His390 is in proximity to the H2 proton of the d-glucitol part of cellobiitol, suggesting that this residue is the most feasible candidate for abstracting the H2 proton in the reaction with the reducing end d-glucose residue (Fig. (C)). The other catalytic His residue, His259, is thought to act as a general acid catalyst, donating a proton to the cis-enediol intermediate. In the reverse reaction, His390 and His259 act as general acid and base catalysts, respectively. Consistent with this observation, His248 of Salmonella enterica AKI (protein YihS), corresponding to His259 of RmCE, is considered to be a general base catalyst in the reaction with d-mannose to generate the cis-enediol intermediate.Citation41) In AKI, His248 is predicted to give the proton, abstracted from H2 of d-mannose, to the C1 of the intermediate to form d-fructose. Considering this prediction, the His residues of C. saccharolyticus and D. turgidum CEs, corresponding to His259 of RmCE, might transfer H2 to C1 of the cis-enediol intermediate to catalyze the isomerization reaction of the reducing end d-glucose or d-mannose residue. However, structural information on these CEs is not sufficient to conclusively determine the mechanism of isomerization catalyzed by CEs, and further analysis is required for better understanding.
Binding of cellobiitol to RmCE indicated the requirement for ring opening, which is a common process as the first step of sugar isomerization/epimerization. In the ring opening, a general acid catalyst donates a proton to the endocyclic oxygen atom, whereas a general base catalyst abstracts a proton from the O1 atom (Fig. (C)). Based on the complexed structures of RmCE and closed substrates (Glcβ1-4Man and epilactose),Citation38) His390 of RmCE is considered to be the sole candidate for the general acid catalyst to donate a proton to O5 of the reducing end sugar residue (Fig. (D)). His200, Glu262, and His390 were suggested as candidates to be a general base catalyst abstracting a proton from O1 of the same residue. The Nε2 atom of His390 was close to both the O5 and O1 atoms of the reducing end of substrates, and thus this residue is predicted to be the most likely candidate for the general acid/base catalyst in the ring opening. The mechanism of ring closure is interpreted as the reverse reaction of ring opening. After the epimerization of the reducing end sugar residue, rotation of the C2–C3 bond may occur again to bring the O1 atom of the reducing end sugar residue close to the O5 atom of the same residue.
In contrast to monosaccharide-specific AGE and AKI, CE is highly specific for disaccharides. Structural analysis of the enzyme–substrate complex of RmCECitation38) clearly identified important amino acid residues for the recognition of the non-reducing end of disaccharides (Fig. (D)). In the complexes of RmCE with Glcβ1-4Man and epilactose, Trp385 on the α11→α12 loop of the enzyme stacks onto the non-reducing end glycosyl residue, and Ser185 and Asp188 on the α5→α6 loop form hydrogen bonds with 4OH and 6OH, respectively. Ser185 and Trp385 are completely conserved in CEs, and many CEs have Asp at the position of RmCE Asp188. In pig AGE,Citation39) an aromatic amino acid residue, Phe377, is situated at the position corresponding to Trp385 of RmCE, but the structure of the α5→α6 loop is very different from that in RmCE and this loop could not interact with the non-reducing end sugar residue of a disaccharide substrate. AKI from S. enterica has α5→α6 and α11→α12 loops with distinct orientations from those in RmCE, and no functionally equivalent amino acid residues are situated at the positions corresponding to Ser185, Asp188, and Trp385 of RmCE.Citation41) Furthermore, superimposition of Glcβ1-4Man bound to RmCE onto the structure of S. enterica AKI showed that the α7→α8 loop of S. enterica AKI clashes with the non-reducing end glucosyl residue, suggesting severe steric hindrance prevents disaccharide binding. These structural differences are presumably the reasons why AGE and AKI lack isomerization/epimerization activity toward disaccharide substrates.
AGE uses monosaccharide substrates harboring an N-acetyl group at the C2 position unlike the substrates of CE and AKI. Although the structure of the enzyme–substrate complex of AGE is still unknown, the structure of RmCE complexed with substrates indicates the structural elements of AGE that might be responsible for binding to the acetamide group of its substrate (Fig. (E)). In the RmCE-substrate complexes, His200, Tyr124, and Asn196 form hydrogen bonds with the 2OH group of the reducing end sugar moiety. Although these amino acid residues are well conserved in AKIs, they are substituted by hydrophobic amino acid residues in AGEs. These three hydrophobic amino acid residues would form a small pocket, which is predicted to accommodate the acetamide group of the AGE substrate.
III. Physiological function of CE in carbohydrate metabolism
Genes neighboring the CE gene suggested the function of CE in carbohydrate metabolism. A putative endo-1,4-β-mannanase (EC 3.2.1.78) gene is encoded near the CE gene in the genome of most CE-producing bacteria for which data are available (Fig. (A)), suggesting that CE is involved in β-mannan metabolism. In B. fragilis, the CE gene (BF0774) is encoded downstream of the ManA gene (BF0771) belonging to glycoside hydrolase family (GH) 26. Cotranscription of BF0771-BF0774 was confirmed,Citation33) and these genes constitute an operon. BF0773 encodes a putative sugar/cation symporter. Biochemical analysis of recombinant protein encoded by BF0772 demonstrated that it catalyzes the specific phosphorolysis of Manβ1-4Glc to α-d-mannose 1-phosphate (Man1P) and d-glucoseCitation33); this enzyme was named 4-O-β-d-mannosyl-d-glucose phosphorylase (EC 2.4.1.281; MGP). Product distribution analysis of ManA protein, predicted to be an extracellular enzyme, showed that this enzyme is a mannan 1,4-β-mannobiosidase (EC 3.2.1.100)Citation43) that specifically produces Manβ1-4Man from β-mannan and β-(1→4)-mannooligosaccharides. Based on these findings, CE is predicted to be involved in the following metabolic pathway (the CE–MGP pathway; Fig. (B)): Manβ1-4Man generated from β-mannan by extracellular ManA is imported into the cytosol through symporter BF0773. CE epimerizes Manβ1-4Man to Manβ1-4Glc, and MGP phosphorolyzes Manβ1-4Glc to Man1P and d-glucose, which are further metabolized through the Embden–Meyerhof–Parnas pathway. A gene organization similar to that in B. fragilis is present in the genome of R. marinus (Fig. (A)). MGP activity of a MGP-like protein, Rmar_2440, was confirmed using a recombinant enzyme preparation.Citation44) The addition of glucomannan to the culture broth of R. marinus clearly induced the production of extracellular β-mannanase and the intracellular enzymes MGP and CE.Citation44) Bacterial growth was also enhanced by supplementation with glucomannan. This observation supports the idea that β-mannanase, CE, and MGP are involved in the metabolism of β-mannan through the CE–MGP pathway.
Fig. 2. The CE–MGP pathway for β-mannan metabolism.
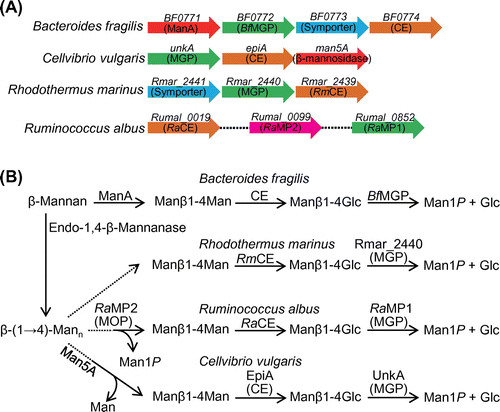
In the genome of R. albus, the CE gene is not found in the gene cluster that was observed in B. fragilis and R. marinus, but two MGP-like genes (Rumal_0099 and Rumal_0852) are present. The deduced amino acid sequences of Rumal_0852 (RaMP1) and Rumal_0099 (RaMP2) are 59% and 27% identical to that of B. fragilis MGP (BfMGP), respectively. MGP activity was detected in cell-free extract of R. albus, and both MGPs were obtained from the cell-free extract, indicating that these two genes are both expressed in R. albus.Citation45) The detailed enzymatic properties of RaMP1 and RaMP2 were investigated using recombinant enzyme samples.Citation45) RaMP1 is a typical MGP, having high selectivity for Manβ1-4Glc, but RaMP2 is not, having markedly higher phosphorolytic activity toward β-(1→4)-mannooligosaccharides longer than Manβ1-4Man compared with its activity toward Manβ1-4Glc. Thus RaMP2 was named β-1,4-mannooligosaccharide phosphorylase (MOP; EC 2.4.1.319). The phosphorolytic activity of RaMP2 toward Manβ1-4Man is low (the kcat/Km is 22-fold lower than that for β-(1→4)-mannotriose), indicating that the physiological function of RaMP2 is to generate Manβ1-4Man from longer β-(1→4)-mannooligosaccharides (Fig. (B)). Manβ1-4Man so generated would be metabolized by RaMP1 and RaCE through the CE–MGP pathway predicted in B. fragilis.
The gene cluster for the CE–MGP pathway in C. vulgaris includes three genes, unkA, epiA, and man5A: unkA encodes a MGP, epiA encodes a CE, and man5A encodes a β-mannosidase that liberates d-mannose from the non-reducing end of β-(1→4)-mannooligosaccharides (Fig. (A)). The α-galactosidase gene aga27A was also found downstream of man5A. Aga27A is considered to split the α-(1→6)-d-galactosyl branch in galactomannan.Citation35) Man5A has much higher hydrolytic activity toward β-(1→4)-mannotriose and β-(1→4)-mannotetraose than Manβ1-4Man.Citation46) This substrate selectivity implies that this enzyme produces Manβ1-4Man for further metabolism by MGP and CE. The difference in enzymes for the degradation of β-(1→4)-mannooligosaccharides in R. albus and C. vulgaris might correlate with the efficiency of ATP production. Phosphorylation using ATP is required for the metabolism of free monosaccharides. In R. albus, MOP is used to avoid the generation of free d-mannose so as to save ATP.
IV. Functions and structures of GH130 mannoside phosphorylases
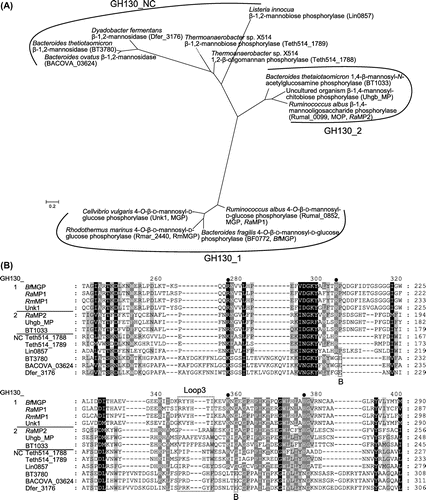
As described above, CE is considered to be involved in β-mannan metabolism along with MGP and MOP. According to sequence-based classification of carbohydrate active enzymes,Citation47) MGP and MOP are members of GH130 together with 1,4-β-mannosyl-N-acetylglucosamine phosphorylase (EC 2.4.1.320),Citation48) β-1,4-mannopyranosyl-chitobiose phosphorylase,Citation49) β-1,2-mannobiose phosphorylase,Citation50) 1,2-β-oligomannan phosphorylaseCitation50), and β-1,2-mannosidase.Citation51,52) Phylogenetic analysis further classified GH130 members into subfamilies GH130_1, GH130_2, and GH130_NC (Fig. (A)): GH130_1 includes MGP; GH130_2 includes MOP, 1,4-β-mannosyl-N-acetylglucosamine phosphorylase, and β-1,4-mannopyranosyl-chitobiose phosphorylase; and GH130_NC includes β-1,2-mannobiose phosphorylase, 1,2-β-oligomannan phosphorylase, and β-1,2-mannosidase.Citation49) 1,4-β-Mannosyl-N-acetylglucosamine phosphorylase and β-1,4-mannopyranosyl-chitobiose phosphorylase, identified in gut Bacteroides, are considered to be involved in the degradation of N-glycan together with GH18 endo-β-N-acetylhexosaminidase (EC 3.2.1.96) and GH92 α-mannosidases (EC 3.2.1.24).Citation48,49) β-1,2-Mannobiose phosphorylase and 1,2-β-oligomannan phosphorylase were predicted to be involved in the biosynthesis of GDP-d-mannose, for which they supply Man1P for the reaction of mannose 1-phosphate guanylyltransferase (EC 2.7.7.22).Citation50) β-1,2-Mannosidase, identified in the intestinal bacterium Bacteroides thetaiotaomicron, is considered to remove the β-(1→2)-mannosyl cap of yeast α-mannan,Citation51) facilitating further degradation of the glycan by the concerted actions of various enzymes.Citation53) β-1,2-Mannosidase was also found in the aerobic bacterium D. fermentans.Citation52)
Fig. 3. Phylogenetic tree and multiple-sequence alignment of GH130 enzymes.
Nakae et al. reported the three-dimensional structure of BfMGP, the first of a GH130 member.Citation54) The catalytic domain of BfMGP is made up of a five-bladed β-propeller fold (Fig. (A)). It has long α-helices at the N- and C-termini, and it forms a homohexamer through hydrophobic contacts via these α-helices. There is no acidic amino acid residue that could directly donate a proton as a general acid catalyst to the scissile glycosidic oxygen in the complex of BfMGP with Manβ1–4Glc. The d-mannosyl residue bound to the −1 subsite adopts a stressed B2,5 boat conformation (Fig. (B)). From these structural features, a unique reaction mechanism was postulated (Fig. (C)): the conserved Asp residue (Asp131 in BfMGP) donates a proton to the 3OH of the d-mannosyl residue in the −1 subsite, and then the 3OH group gives a proton to the glycosidic oxygen to split the β-mannosidic linkage (i.e. there is a proton relay mechanism). This reaction mechanism is definitely different from that of the inverting phosphorylases categorized in GH families, because the general acid catalyst of such inverting phosphorylases directly donates a proton to the scissile glycosidic oxygen.Citation55–57) The 3OH group of the d-mannosyl residue in the −1 subsite is situated in an intermediate position between the catalytic Asp and the scissile glycosidic oxygen in other GH130 enzymes,Citation58,59) and these structural data support the proton relay reaction mechanism postulated by Nakae et al.Citation54) although β-mannosidase is thought to transfer the proton from the general acid catalyst to the glycosidic oxygen via solvent.Citation51)
Fig. 4. Structure–function relationship of GH130_1 and GH130_2 mannoside phosphorylases.
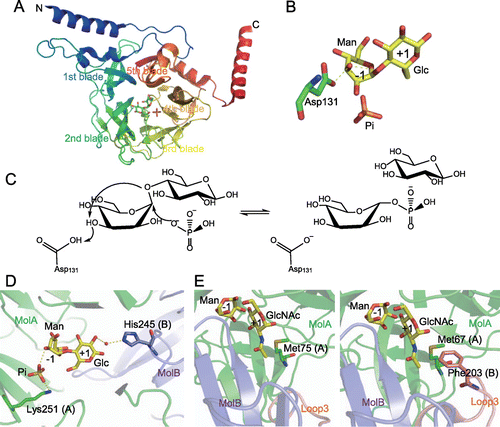
Structural analysis of RaMP1 revealed that, unlike BfMGP, this enzyme forms a homotrimer.Citation59) This difference in the quaternary structure is thought to be due to the substitution of hydrophobic amino acid residues found in the N-terminal α-helix of BfMGP with hydrophilic residues in RaMP1. Furthermore, in this structural analysis, it was found that a loop, named Loop3, from the adjacent monomer contributed to the formation of the +1 subsite, and restricted the formation of further “+” subsites, which is an important factor in the disaccharide specificity of the enzyme. His245 on Loop3, which is conserved only in GH130_1 subfamily enzymes (Fig. (B)), forms a hydrogen bond with the 2OH of the d-glucose residue in the +1 subsite of the neighboring monomer via a water molecule (Fig. (D)), and it was shown to be important for catalytic activity through site-directed mutation. His245 is involved in a hydrogen bond network including this water, Manβ1-4Glc, inorganic phosphate, and Lys251 binding to the inorganic phosphate. As the substitution of His245 with Ala significantly decreased the affinity for Man1P in the reverse phosphorolysis, His245 is also important for substrate binding in the −1 subsite. His245 might contribute to substrate binding to the −1 subsite by stabilizing the hydrogen bond network.
Compared with disaccharide-specific GH130_1 MGPs, GH130_2 enzymes have wide-open substrate-binding sites.Citation58,59) GH130_2 enzymes accept long-chain substrates with a degree of polymerization ≥3, and this open substrate-binding site is necessary to accommodate substrates longer than disaccharides. In contrast to GH130_1, subfamily GH130_2 contains mannoside phosphorylases with different substrate specificities. MOP strongly prefers a d-mannose moiety to a N-acetyl-d-glucosamine moiety at the +1 subsite, whereas 1,4-β-mannosyl-N-acetylglucosamine phosphorylase and β-1,4-mannopyranosyl-chitobiose phosphorylase have high selectivity for an N-acetyl-d-glucosamine moiety. Structural analysis of homohexameric Uhgb_MP, a β-1,4-mannopyranosyl-chitobiose phosphorylase from an uncultured gut bacterium, suggested that Met67 and Phe203 (situated on Loop3 of the adjacent molecule) side chains form a hydrophobic pocket interacting with the methyl group of the N-acetyl-d-glucosamine moiety in the +1 subsite (Fig. (E)).Citation58) RaMP2, a MOP from R. albus, has Met75 at the position corresponding to Met67 of Uhgb_MP, but its Loop3 is shorter than that of Uhgb_MP, and it has no hydrophobic interaction with the acetamide group.Citation59) This structural feature might determine the substrate specificity of GH130_2 enzymes.
β-Mannosidases, catalyzing hydrolysis of a non-reducing end β-(1→2)-mannosidic linkage with net inversion, were recently identified among GH130_NC members.Citation51,52) They lack basic amino acid residues that are highly conserved in GH130 phosphorylases and responsible for positioning phosphate (Fig. (B)). Inverting glycosidases catalyze hydrolysis through a single-displacement mechanism, in which a general acid catalyst donates a proton to the glycosidic oxygen, and a water molecule, activated by a general base catalyst, performs nucleophilic attack on the anomeric carbon. In these β-mannosidases, two Glu residues, conserved only in β-mannosidases (Fig. (B)), are predicted to coordinate a water molecule for the nucleophilic attack. Substitutions of both these Glu residues completely abolished the catalytic activity,Citation51,52) and thus they were considered to act as the general base catalyst.
V. Application of CE for production of functional oligosaccharides
Epilactose is present at a very low level in heated milk and alkaline-treated lactose.Citation60,61) Its physiological function was unclear for a long time because of difficulty in preparing this compound. Miyasato and Ajisaka reported enzymatic production of epilactose by transgalactosylation catalyzed by β-galactosidase.Citation62) In this reaction system, a d-galactosyl residue is transferred from a synthetic substrate, p-nitrophenyl β-d-galactopyranoside, to acceptor d-mannose. However, it is very difficult to synthesize epilactose on a >10-g scale by this method. Finding the epimerization activity of CE toward readily available lactoseCitation20) made large-scale preparation of epilactose possible; in this reaction, approximately 30% of the lactose is converted to epilactose.Citation22,23) A continuous reaction system using CE immobilized on anion-exchange resin effectively reduces the amount of enzyme required for epilactose synthesis.Citation63) The epilactose generated can be purified from the reaction mixture in several steps to approximately 90% purity; these steps include crystallization of residual lactose, hydrolysis of lactose by β-galactosidase, degradation of the resulting monosaccharides by yeast, and column chromatography using Na-form cation-exchange resin.Citation22) All these steps can be scaled up, thus this purification procedure can be used industrially.
In vitro experiments revealed that epilactose is very stable with respect to intestinal digestive enzymes and enhances the growth of some bifidobacterial strains.Citation20) Stimulation of growth of bifidobacteria was confirmed in animal experiments using rats.Citation64) Oral administration of epilactose inhibited conversion of primary bile acids to secondary bile acids,Citation64) which are considered to be risk factors for colon cancer, and stimulated mineral absorption.Citation65) Enhanced mineral absorption is predicted to be caused by lowering the pH through the production of short-chain fatty acids by intestinal bacteria proliferated by epilactose. Furthermore, Suzuki et al. clearly demonstrated that epilactose increased paracellular Ca2+ absorption in the small intestine by inducing the phosphorylation of myosin regulatory light chain by myosin light chain kinase and Rho-associated kinase.Citation66) As a result of enhanced mineral absorption following the ingestion of epilactose, postgastrectomy osteopenia and anemia were improved in rat experiments,Citation67) suggesting that epilactose is a superior candidate functional foodstuff to prevent osteoporosis and anemia. Furthermore, recently, we reported that epilactose effectively prevents high-fat diet-induced obesity in mice.Citation68) Epilactose feeding increased the expression of uncoupling protein 1, which is involved in dissipation of energy, in skeletal muscles and brown adipose tissue, leading to an increase in whole-body energy expenditure. The expression of uncoupling protein 1 is enhanced by propionic acid, which is generated by intestinal microflora that use epilactose as a carbon source. Thus epilactose has great potential to prevent obesity and related diseases, recognized as major health problems, particularly in developed countries.
The isomerization activity of CE from C. saccharolyticus is used in the efficient enzymatic production of lactulose,Citation34) a commercially available prebiotic oligosaccharide. Although epilactose is also generated as a byproduct (15% yield), 58% yield of lactulose was obtained in the reaction of 700 g/L lactose substrate. As lactulose is currently produced by alkaline isomerization of lactose, the production process includes complicated purification and desalting steps. An environmentally friendly synthetic method using a biocatalyst is preferable to the conventional chemical reaction.
CEs are fully active in milk, and thus direct production of epilactose and lactulose has been established.Citation26,69,70) Highly purified lactose is required for the production of galactooligosaccharide and lactulose by chemical reaction, because the substrate concentration must be high enough for efficient transgalactosylation for galactooligosaccharide production and the purity of the substrate must be high for chemical isomerization of lactose to avoid side reactions. Thus, the direct production of prebiotic oligosaccharides using CEs in milk is a very attractive alternative approach to easily convert milk to functional dairy products.
Funding
This work was supported in part by a Grant-in-Aid for Young Scientists from the Ministry of Education, Culture, Sports, Science, and Technology of Japan [grant number 24780091], [grant number 26850059].
Acknowledgements
I am very grateful to Professor Emeritus Hirokazu Matsui of Hokkaido University for continuous encouragement. I sincerely thank Professor Haruhide Mori, Professor Atsuo Kimura, and Dr Masayuki Okuyama of the Research Faculty of Agriculture, Hokkaido University, for their fruitful discussions. I was only able to carry out structural analysis of CEs and GH130 enzymes in collaboration with Professor Min Yao, Dr Koji Kato, Dr Takaaki Fujiwara, Dr Ye Yuxin, and the other members of the Laboratory of X-ray Structural Biology of the Faculty of Advanced Bioscience, Hokkaido University. NMR analyses of oligosaccharides generated by the enzymes were performed in cooperation with Dr Eri Fukushi of the GC–MS & NMR Laboratory, Research Faculty of Agriculture, Hokkaido University. MS and amino acid analyses of the enzyme reaction products and the enzymes prepared, respectively, were carried out by Mr Tomohiro Hirose and Ms Nozomi Takeda of the Global Facility Center, Hokkaido University. Physiological functions of epilactose were analyzed in collaboration with Dr Takuya Suzuki of Hiroshima University. Man1P used for the biochemical analysis of GH130 enzymes was kindly supplied by Dr Motomitsu Kitaoka and Dr Mamoru Nishimoto of the National Food Research Institute, National Agriculture and Food Research Organization. I thank Dr Teruyo Kato-Ojima of Nagoya University for much assistance in the biochemical analysis of CEs and preparation of epilactose. I thank all of my co-workers in the Laboratory of Biochemistry, Research Faculty of Agriculture, Hokkaido University, for their cooperation. Part of these studies was supported by a Grant-in-Aid for Young Scientists from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Disclosure statement
No potential conflict of interest was reported by the author.
Notes
This review was written in response to the author’s receipt of the JSBBA Award for Young Scientists in 2015.
Abbreviations: AGE, N-acylglucosamine 2-epimerase; AKI, aldose-ketose isomerase; BfMGP, Bacteroides fragilis 4-O-β-d-mannosyl-d-glucose phosphorylase; CE, cellobiose 2-epimerase; GH, glycoside hydrolase family; Glcβ1-4Man, β-d-glucopyranosyl-(1→4)-d-mannose; Manβ1-4Glc, β-d-mannopyranosyl-(1→4)-d-glucose; Manβ1-4Man, β-(1→4)-mannobiose; MGP, 4-O-β-d-mannosyl-d-glucose phosphorylase; MOP, β-1,4-mannooligosaccharide phosphorylase; Man1P, α-d-mannose 1-phosphate; RaCE, Ruminococcus albus CE; RaMP1, R. albus MGP; RaMP2, R. albus MOP; RmCE, Rhodothermus marinus CE; Uhgb_MP, β-1,4-mannopyranosyl-chitobiose phosphorylase from an uncultured gut bacterium.
References
- Gibson GR, Beatty ER, Wang X, et al. Selective stimulation of bifidobacteria in the human colon by oligofructose and inulin. Gastroenterology. 1995;108:975–982.10.1016/0016-5085(95)90192-2
- Tanaka R, Takayama H, Morotomi M, et al. Effects of administration of TOS and Bifidobacterium breve 4006 on the Human Fecal Flora. Bifidobact. Microflora. 1983;2:17–24.10.12938/bifidus1982.2.1_17
- Bouhnik Y, Attar A, Joly FA, et al. Lactulose ingestion increases faecal bifidobacterial counts: a randomised double-blind study in healthy humans. Eur. J. Clin. Nutr. 2004;58:462–466.10.1038/sj.ejcn.1601829
- Kaneko T, Kohmoto T, Kikuchi H, et al. Effects of isomaltooligosaccharides with different degrees of polymerization on human fecal bifidobacteria. Biosci. Biotechnol. Biochem. 1994;58:2288–2290.10.1271/bbb.58.2288
- Murosaki S, Muroyama K, Yamamoto Y, et al. Immunopotentiating activity of nigerooligosaccharides for the T helper 1-like immune response in mice. Biosci. Biotechnol. Biochem. 1999;63:373–378.10.1271/bbb.63.373
- Kovacs-Nolan J, Kanatani H, Nakamura A, et al. β-1,4-Mannobiose stimulates innate immune responses and induces TLR4-dependent activation of mouse macrophages but reduces severity of inflammation during endotoxemia in mice. J. Nutr. 2013;143:384–391.10.3945/jn.112.167866
- Yamanaka K. Purification, crystallization and properties of the d-xylose isomerase from Lactobacillus brevis. Biochim. Biophys. Acta. 1968;151:670–680.10.1016/0005-2744(68)90015-6
- Shamanna DK, Sanderson KE. Uptake and catabolism of d-xylose in Salmonella typhimurium LT2. J. Bacteriol. 1979;139:64–70.
- Bhosale SH, Rao MB, Deshpande VV. Molecular and industrial aspects of glucose isomerase. Microbiol. Rev. 1996;60:280–300.
- Kuyper M, Winkler AA, van Dijken JP, et al. Minimal metabolic engineering of Saccharomyces cerevisiae for efficient anaerobic xylose fermentation: a proof of principle. FEMS Yeast Res. 2004;4:655–664.10.1016/j.femsyr.2004.01.003
- Itoh H, Okaya H, Khan AR, et al. Purification and characterization of d-tagatose 3-epimerase from Pseudomonas sp. ST-24. Biosci. Biotechnol. Biochem. 1994;58:2168–2171.10.1271/bbb.58.2168
- Kim HJ, Hyun EK, Kim YS, et al. Characterization of an Agrobacterium tumefaciens d-psicose 3-epimerase that converts d-fructose to d-psicose. Appl. Environ. Microbiol. 2006;72:981–985.10.1128/AEM.72.2.981-985.2006
- Takeshita K, Suga A, Takada G, et al. Mass production of d-psicose from d-fructose by a continuous bioreactor system using immobilized d-tagatose 3-epimerase. J. Biosci. Bioeng. 2000;90:453–455.10.1016/S1389-1723(01)80018-9
- Iida T, Hayashi N, Yamada T, et al. Failure of d-psicose absorbed in the small intestine to metabolize into energy and its low large intestinal fermentability in humans. Metabolism. 2010;59:206–214.10.1016/j.metabol.2009.07.018
- Matsuo T, Izumori K. d-Psicose inhibits intestinal α-glucosidase and suppresses the glycemic response after ingestion of carbohydrates in rats. J. Clin. Biochem. Nutr. 2009;45:202–206.10.3164/jcbn.09-36
- Ochiai M, Nakanishi Y, Yamada T, et al. Inhibition by dietary d-psicose of body fat accumulation in adult rats fed a high-sucrose diet. Biosci. Biotechnol. Biochem. 2013;77:1123–1126.10.1271/bbb.130019
- Ochiai M, Onishi K, Yamada T, et al. d-Psicose increases energy expenditure and decreases body fat accumulation in rats fed a high-sucrose diet. Int. J. Food Sci. Nutr. 2014;65:245–250.10.3109/09637486.2013.845653
- Tyler TR, Leatherwood JM. Epimerization of disaccharides by enzyme preparations from Ruminococcus albus. Arch. Biochem. Biophys. 1967;119:363–367.10.1016/0003-9861(67)90466-3
- Ito S, Hamada S, Yamaguchi K, et al. Cloning and sequencing of the cellobiose 2-epimerase gene from an obligatory anaerobe, Ruminococcus albus. Biochem. Biophys. Res. Commun. 2007;360:640–645.10.1016/j.bbrc.2007.06.091
- Ito S, Taguchi H, Hamada S, et al. Enzymatic properties of cellobiose 2-epimerase from Ruminococcus albus and the synthesis of rare oligosaccharides by the enzyme. Appl. Microbiol. Biotechnol. 2008;79:433–441.10.1007/s00253-008-1449-7
- Jaito N, Saburi W, Muto H, et al. Colorimetric quantification of β-(1→4)-mannobiose and 4-O-β-d-mannosyl-d-glucose. J. Appl. Glycosci. 2014;61:117–119.10.5458/jag.jag.JAG-2014_007
- Saburi W, Yamamoto T, Taguchi H, et al. Practical preparation of epilactose produced with cellobiose 2-epimerase from Ruminococcus albus NE1. Biosci. Biotechnol. Biochem. 2010;74:1736–1737.10.1271/bbb.100353
- Ojima T, Saburi W, Sato H, et al. Biochemical characterization of a thermophilic cellobiose 2-epimerase from a thermohalophilic bacterium, Rhodothermus marinus JCM9785. Biosci. Biotechnol. Biochem. 2011;75:2162–2168.10.1271/bbb.110456
- Senoura T, Taguchi H, Ito S, et al. Identification of the cellobiose 2-epimerase gene in the genome of Bacteroides fragilis NCTC 9343. Biosci. Biotechnol. Biochem. 2009;73:400–406.10.1271/bbb.80691
- Park CS, Kim JE, Choi JG, et al. Characterization of a recombinant cellobiose 2-epimerase from Caldicellulosiruptor saccharolyticus and its application in the production of mannose from glucose. Appl. Microbiol. Biotechnol. 2011;92:1187–1196.10.1007/s00253-011-3403-3
- Krewinkel M, Kaiser J, Merz M, et al. Novel cellobiose 2-epimerases for the production of epilactose from milk ultrafiltrate containing lactose. J. Dairy Sci. 2015;98:3665–3678.10.3168/jds.2015-9411
- Saburi W, Tanaka Y, Muto H, et al. Functional reassignment of Cellvibrio vulgaris EpiA to cellobiose 2-epimerase and an evaluation of the biochemical functions of the 4-O-β-d-mannosyl-d-glucose phosphorylase-like protein, UnkA. Biosci. Biotechnol. Biochem. 2015;79:969–977.10.1080/09168451.2015.1012146
- Kim JE, Kim YS, Kang LW, et al. Characterization of a recombinant cellobiose 2-epimerase from Dictyoglomus turgidum that epimerizes and isomerizes β-1,4- and α-1,4-gluco-oligosaccharides. Biotechnol. Lett. 2012;34:2061–2068.10.1007/s10529-012-0999-z
- Ojima T, Saburi W, Yamamoto T, et al. Identification and characterization of cellobiose 2-epimerase from various aerobes. Biosci. Biotechnol. Biochem. 2013;77:189–193.10.1271/bbb.120742
- Taguchi H, Senoura T, Hamada S, et al. Cloning and sequencing of the gene for cellobiose 2-epimerase from a ruminal strain of Eubacterium cellulosolvens. FEMS Microbiol. Lett. 2008;287:34–40.10.1111/fml.2008.287.issue-1
- Park CS, Kim JE, Lee SH, et al. Characterization of a recombinant mannobiose 2-epimerase from Spirochaeta thermophila that is suggested to be a cellobiose 2-epimerase. Biotechnol. Lett. 2013;35:1873–1880.10.1007/s10529-013-1267-6
- Wasaki J, Taguchi H, Senoura T, et al. Identification and distribution of cellobiose 2-epimerase genes by a PCR-based metagenomic approach. Appl. Microbiol. Biotechnol. 2015;99:4287–4295.10.1007/s00253-014-6265-7
- Senoura T, Ito S, Taguchi H, et al. New microbial mannan catabolic pathway that involves a novel mannosylglucose phosphorylase. Biochem. Biophys Res. Commun. 2011;408:701–706.10.1016/j.bbrc.2011.04.095
- Kim YS, Oh DK. Lactulose production from lactose as a single substrate by a thermostable cellobiose 2-epimerase from Caldicellulosiruptor saccharolyticus. Bioresour. Technol. 2012;104:668–672.10.1016/j.biortech.2011.11.016
- Centeno MS, Guerreiro CI, Dias FM, et al. Galactomannan hydrolysis and mannose metabolism in Cellvibrio mixtus. FEMS Microbiol. Lett. 2006;261:123–132.10.1111/fml.2006.261.issue-1
- Amein M, Leatherwood JM. Mechanism of cellobiose epimerase. Biochem. Biophys. Res. Commun. 1969;36:223–227.10.1016/0006-291X(69)90318-0
- Fujiwara T, Saburi W, Inoue S, et al. Crystal structure of Ruminococcus albus cellobiose 2-epimerase: structural insights into epimerization of unmodified sugar. FEBS Lett. 2013;587:840–846.10.1016/j.febslet.2013.02.007
- Fujiwara T, Saburi W, Matsui H, et al. Structural insights into the epimerization of β-1,4-linked oligosaccharides catalyzed by cellobiose 2-epimerase, the sole enzyme epimerizing non-anomeric hydroxyl groups of unmodified sugars. J. Biol. Chem. 2014;289:3405–3415.10.1074/jbc.M113.531251
- Itoh T, Mikami B, Maru I, et al. Crystal structure of N-acyl-d-glucosamine 2-epimerase from porcine kidney at 2.0 Å resolution. J. Mol. Biol. 2000;303:733–744.10.1006/jmbi.2000.4188
- Lee YC, Wu HM, Chang YN, et al. The central cavity from the (α/α)6 barrel structure of Anabaena sp. CH1 N-acetyl-d-glucosamine 2-epimerase contains two key histidine residues for reversible conversion. J. Mol. Biol. 2007;367:895–908.10.1016/j.jmb.2006.11.001
- Itoh T, Mikami B, Hashimoto W, et al. Crystal structure of YihS in complex with d-mannose: structural annotation of Escherichia coli and Salmonella enterica yihS-encoded proteins to an aldose-ketose isomerase. J. Mol. Biol. 2008;377:1443–1459.10.1016/j.jmb.2008.01.090
- Ito S, Hamada S, Ito H, et al. Site-directed mutagenesis of possible catalytic residues of cellobiose 2-epimerase from Ruminococcus albus. Biotechnol. Lett. 2009;31:1065–1071.10.1007/s10529-009-9979-3
- Kawaguchi K, Senoura T, Ito S, et al. The mannobiose-forming exo-mannanase involved in a new mannan catabolic pathway in Bacteroides fragilis. Arch. Microbiol. 2014;196:17–23.10.1007/s00203-013-0938-y
- Jaito N, Saburi W, Odaka R, et al. Characterization of a thermophilic 4-O-β-d-mannosyl-d-glucose phosphorylase from Rhodothermus marinus. Biosci. Biotechnol. Biochem. 2014;78:263–270.10.1080/09168451.2014.882760
- Kawahara R, Saburi W, Odaka R, et al. Metabolic mechanism of mannan in a ruminal bacterium, Ruminococcus albus, involving two mannoside phosphorylases and cellobiose 2-epimerase: discovery of a new carbohydrate phosphorylase, β-1,4-mannooligosaccharide phosphorylase. J. Biol. Chem. 2012;287:42389–42399.10.1074/jbc.M112.390336
- Dias FM, Vincent F, Pell G, et al. Insights into the molecular determinants of substrate specificity in glycoside hydrolase family 5 revealed by the crystal structure and kinetics of Cellvibrio mixtus mannosidase 5A. J. Biol. Chem. 2004;279:25517–25526.10.1074/jbc.M401647200
- Lombard V, Golaconda Ramulu H, Drula E, et al. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42:D490–D495.10.1093/nar/gkt1178
- Nihira T, Suzuki E, Kitaoka M, et al. Discovery of β-1,4-d-mannosyl-N-acetyl-d-glucosamine phosphorylase involved in the metabolism of N-glycans. J. Biol. Chem. 2013;288:27366–27374.10.1074/jbc.M113.469080
- Ladevèze S, Tarquis L, Cecchini DA, et al. Role of glycoside phosphorylases in mannose foraging by human gut bacteria. J. Biol. Chem. 2013;288:32370–32383.10.1074/jbc.M113.483628
- Chiku K, Nihira T, Suzuki E, et al. Discovery of two β-1,2-mannoside phosphorylases showing different chain-length specificities from thermoanaerobacter sp. X-514. PLoS One. 2014;9:e114882.10.1371/journal.pone.0114882
- Cuskin F, Baslé A, Ladevèze S, et al. The GH130 family of mannoside phosphorylases contains glycoside hydrolases that target β-1,2-mannosidic linkages in Candida mannan. J. Biol. Chem. 2015;290:25023–25033.10.1074/jbc.M115.681460
- Nihira T, Chiku K, Suzuki E, et al. An inverting β-1,2-mannosidase belonging to glycoside hydrolase family 130 from Dyadobacter fermentans. FEBS Lett. 2015;589:3604–3610.10.1016/j.febslet.2015.10.008
- Cuskin F, Lowe EC, Temple MJ, et al. Human gut Bacteroidetes can utilize yeast mannan through a selfish mechanism. Nature. 2015;517:165–169.10.1038/nature13995
- Nakae S, Ito S, Higa M, et al. Structure of novel enzyme in mannan biodegradation process 4-O-β-d-mannosyl-d-glucose phosphorylase MGP. J. Mol. Biol. 2013;425:4468–4478.10.1016/j.jmb.2013.08.002
- Egloff MP, Uppenberg J, Haalck L, et al. Crystal structure of maltose phosphorylase from Lactobacillus brevis: unexpected evolutionary relationship with glucoamylase. Structure. 2001;9:689–697.10.1016/S0969-2126(01)00626-8
- Hidaka M, Honda Y, Kitaoka M, et al. Chitobiose phosphorylase from Vibrio proteolyticus, a member of glycosyl transferase family 36, has a clan GH-L-like (α/α)6 barrel fold. Structure. 2004;12:937–947.10.1016/j.str.2004.03.027
- Hidaka M, Nishimoto M, Kitaoka M, et al. The crystal structure of galacto-N-biose/lacto-N-biose I phosphorylase: a large deformation of a TIM barrel scaffold. J. Biol. Chem. 2009;284:7273–7283.10.1074/jbc.M808525200
- Ladevèze S, Cioci G, Roblin P, et al. Structural bases for N-glycan processing by mannoside phosphorylase. Acta Crystallogr. D Biol. Crystallogr. 2015;71:1335–1346.10.1107/S1399004715006604
- Ye X, Saburi W, Odaka R, et al. Structural insights into the difference in substrate recognition of two mannoside phosphorylases from two GH130 subfamilies. FEBS Lett. Forthcoming.
- Martinez-Castro I, Olano A. Influence of thermal processing on carbohydrate composition of milk. Formation of epilactose. Milchwissenschaft. 1980;35:5–8.
- Olano A, Martinez-Castro I. Formation of lactulose and epilactose from lactose in basic media. A quantitative study. Milchwissenschaft. 1981;36:533–536.
- Miyasato M, Ajisaka K. Regioselectivity in β-galactosidase-catalyzed transglycosylation for the enzymatic assembly of d-galactosyl-d-mannose. Biosci. Biotechnol. Biochem. 2004;68:2086–2090.10.1271/bbb.68.2086
- Sato H, Saburi W, Ojima T, et al. Immobilization of a thermostable cellobiose 2-epimerase from Rhodothermus marinus JCM9785 and continuous production of epilactose. Biosci. Biotechnol. Biochem. 2012;76:1584–1587.10.1271/bbb.120284
- Watanabe J, Nishimukai M, Taguchi H, et al. Prebiotic properties of epilactose. J. Dairy Sci. 2008;91:4518–4526.10.3168/jds.2008-1367
- Nishimukai M, Watanabe J, Taguchi H, et al. Effects of epilactose on calcium absorption and serum lipid metabolism in rats. J. Agric. Food Chem. 2008;56:10340–10345.10.1021/jf801556m
- Suzuki T, Nishimukai M, Takechi M, et al. The Nondigestible disaccharide epilactose increases paracellular Ca absorption via rho-associated kinase- and myosin light chain kinase-dependent mechanisms in rat small intestines. J. Agric. Food Chem. 2010;58:1927–1932.10.1021/jf9035063
- Suzuki T, Nishimukai M, Shinoki A, et al. Ingestion of epilactose, a non-digestible disaccharide, improves postgastrectomy osteopenia and anemia in rats through the promotion of intestinal calcium and iron absorption. J. Agric. Food Chem. 2010;58:10787–10792.10.1021/jf102563y
- Murakami Y, Ojima-Kato T, Saburi W, et al. Supplemental epilactose prevents metabolic disorders through uncoupling protein-1 induction in the skeletal muscle of mice fed high-fat diets. Br. J. Nutr. 2015;114:1774–1783.10.1017/S0007114515003505
- Krewinkel M, Gosch M, Rentschler E, et al. Epilactose production by 2 cellobiose 2-epimerases in natural milk. J. Dairy Sci. 2014;97:155–161.10.3168/jds.2013-7389
- Rentschler E, Schuh K, Krewinkel M, et al. Enzymatic production of lactulose and epilactose in milk. J. Dairy Sci. 2015;98:6767–6775.10.3168/jds.2015-9900
