Abstract
We analyzed the role of the nitrate transporter-encoding gene (nrtA) of Aspergillus oryzae by gene disruption. Southern hybridization analysis indicated that homologous recombination occurred at the resident nrtA locus. Real-time PCR showed that the nrtA gene was strongly inducible by NaNO3. The nrtA disruptant did not exhibit normal growth when nitrate was available as the sole nitrogen source. These results indicate that NrtA is essential for nitrate uptake in A. oryzae. Kojic acid (KA) production was inhibited by the addition of a small amount of sodium nitrate. The nrtA-disrupted strain was deficient in the uptake of nitrate. As a result, KA production in this strain was not considerably affected by the presence of nitrate.
Graphical abstract
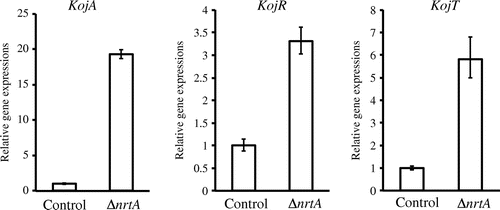
The expression levels of the KA biosynthesis genes (kojA, kojR, and kojT) increased markedly in the nrtA-disrupted strain in the presence of nitrate.
Key words:
Melanin, the main determinant of skin color, confers protection against ultraviolet (UV) radiation. Melanogenesis is a complex process involving the activity of tyrosinase and tyrosinase-related proteins. Tyrosinase plays an important role in regulating melanin formation, during which l-tyrosine is hydroxylated to l-dihydroxyphenylalanine (l-DOPA), and l-DOPA is oxidized into o-quinone.Citation1) The use of tyrosinase inhibitors is increasingly common in the cosmetics industry because of the skin-whitening effects of these compounds.Citation2)
Kojic acid (KA), which inhibits tyrosinase by chelating copper ions required for the active site of the enzyme, is used as a skin-lightening agent in the cosmetics industry.Citation3) KA is a major secondary metabolite produced by aerobic fermentation with Aspergillus species such as Aspergillus oryzae, Aspergillus flavus, and Aspergillus tamarii.Citation4,5) Despite extensive studies, neither the biosynthetic pathway nor the genes involved in KA production have been identified since KA was first isolated over a 100 years ago. Recently, several regions of the KA-encoding gene in A. oryzae were identified by microarray and gene disruption analysis. A transcription factor gene (kojR), an enzyme gene (kojA), and a transporter gene (kojT) were found to constitute the KA biosynthetic cluster. The kojR gene encodes a fungal-specific Zn(II)2Cy6 transcription factor that regulates the transcription of kojA and kojT. KA production is dependent on the concentration of the nitrogen source. The addition of a small amount of sodium nitrate to standard medium resulted in a complete loss of KA production, indicating that this compound functions as an inhibitor of KA production.Citation6,7)
Nitrate is a major nitrogen source for most micro-organisms. The uptake of nitrate by cells is an active process that occurs against a nitrate gradient and is facilitated by a nitrate transporter. The nitrate transporter family, which consists of both prokaryotic and eukaryotic members, is one of the 17 families of transporters that comprise the major facilitator superfamily (MFS). Members of the MFS family are typically composed of 500–600 amino acids, with 12 hydrophobic transmembrane domains in α-helical conformation passing through the membrane and connected by hydrophilic loops.Citation8,9) Nitrate uptake has been well studied in the fungus Aspergillus nidulans. In A. nidulans, two nitrate transporters, NrtA (formerly CrnA) and NrtB, which share 61% sequence identity, have been identified.Citation10,11) Mutant strains of A. nidulans lacking both nrtA and nrtB have been shown to be unable to grow in standard minimal medium containing 10 mM of NO3−.Citation12,13)
We assume that the gene-disrupted strain lacking the nitrate transporter gene is able to produce KA in the presence of nitrate. In this paper, we analyzed the function of the nitrate transporter in A. oryzae. Furthermore, we produced an nrtA disruption mutant lacking the nitrate transporter and evaluated KA production by the nrtA-disrupted strain in the presence of nitrate.
Materials and methods
Strains and culture conditions
A. oryzae RIB40 was obtained from the National Research Institute of Brewing, Japan (http://www.nrib.go.jp/ken/asp/strain.html). For genetic engineering, an A. oryzae strain with the genotype ligD::ptrA (pyrG-) was used as the host strain for transformation.Citation14) The control strain for these experiments was transformed with pyrG only. The nrtA-disrupted mutant and control strains were grown on yeast peptone dextrose (YPD) medium (Becton, Dickinson and Company, Sparks, MD) at 30 °C for 24 h. DNA was extracted using the Wizard® Genomic DNA Purification Kit (Promega, Madison, WI) according to the manufacturer’s instructions.
Conidia of A. oryzae grown on Czapek-Dox (CD) agar medium (Becton, Dickinson and Company) were harvested and stocked until used. The stocked conidia were spread on Difco PD agar medium (Becton, Dickinson and Company), and generated conidia were harvested and used for successive submerged culture to produce KA. Typically, 105 conidia/ml was inoculated into the liquid medium. The standard medium for KA production contained 0.25% yeast extract, 0.1% K2HPO4, 0.05% MgSO4–7H2O, and 10% glucose. When approximately 105 conidia/ml of A. oryzae RIB40 were inoculated into standard liquid medium, KA secretion was detected within 6 days after inoculation.Citation15,16)
Expression analysis by real-time PCR
To analyze the expression of nrtA using real-time PCR, we isolated total RNA from the control and disrupted strains grown in YPD medium at 30 °C for 24 h. The main culture was then established for 6 h at 30 °C in minimal medium (2% glucose, 0.3% KH2PO4, 0.1% KCl, and 0.1% MgSO4) containing 100 mM NaNO3 or 100 mM (NH4)2SO4. Total RNA was extracted using RNAiso Plus (TaKaRa, Shiga, Japan) according to the manufacturer’s instructions. mRNA was purified using an Oligotex® dT30 (super) mRNA Purification Kit (TaKaRa) following the manufacturer’s protocol. Reverse transcription and real-time PCR were performed as previously describedCitation17) using the SuperScript III Platinum Two-Step qRT-PCR Kit with SYBR Green® (Invitrogen, Carlsbad, CA). Relative quantification was performed using the 2−ΔΔCt technique (ABI User Bulletin 2). The sequences (5′→3′) of the nrtA primers were CTGCTGTGTGGTTCTATTCCAACT (forward) and GACAAAGAAGCGTAAGGCAATCA (reverse).
KA biosynthesis gene (KojA, KojR, and KojT) expression levels in the nrtA-disrupted strain were analyzed using real-time PCR. We isolated total RNA and mRNA from the control and the nrtA-disrupted strain grown in the standard medium for KA production, with 0.3% sodium nitrate supplementation, at 30 °C for 3 days. The primer sequences (5′→3′) were as follows: ACACAAACGAGCCCCTTCAG (KojA-F), CCTCGTGACGGTCGAATGA (KojA-R), CAACTCAGGCACCGCTTTC (KojR-F), TCCAGCTAAACCCGTACACCTT (KojR-R), TTTGGAGGATGCCGACGAT (KojT-F), and TTCCTGGCGTGGAGTATGG (KojT-R).
Histone H1 was used as an endogenous control. The sequences (5′→3′) of the histone H1 primers were GACAACATCCAGGGTATCACTAAGC (forward) and CGGGTCTCCTCGTAGATCATGGCAG (reverse).
Construction of transformant strains
∆nrtA was created as described by Tamano.Citation18) The nrtA gene disruption cassette was generated by fusion PCR using an Expand High Fidelity PCR System (Roche Diagnostics, Mannheim, Germany). The 5′- and 3′-arms of nrtA were amplified from genomic DNA using the primers LU/LL (for the 5′-arm) and RU/RL (for the 3′-arm). pyrG was amplified using primer pair PU/PL. Amplified fragments were purified using the Wizard Gel extraction kit (Promega, Madison, WI). The 5′-flanking:pyrG:3′-flanking amplicon (1:3:1 M ratio) was used as a template. The PCR products were used for a second round of PCR with the primer pair LU/RL to fuse the 5′ and 3′ regions of the target gene at each end of the pyrG gene. The primer sequences (5′→3′) were as follows: CATCGAAAGTCTAACCAG (LU), CACAGGGTACGTCTGTTGTCAGTTGGAATAGAACCAC (LL), GTGGTTCTATTCCAACTGACAACAGACGTACCCTGTGATGTTC (PU), AAGCTGGCGAGCAAGAAACTGCACCTCAGAAGAAAAGGATG (PL), TTCTTCTGAGGTGCAGTTTCTTGCTCGCCAGCTTCGT (RU), and CCAGAGTGAGCGTGCGTAGT (RL). The amplified fragment was purified using the Wizard Gel extraction kit and subsequently used for transformation.
Fungal transformation was performed as described by Takahashi.Citation19) The uridine-prototrophic transformants were subjected to two consecutive rounds of single sporing. Southern blotting of genomic DNA was performed following digestion with HindIII. The DNA fragment containing the 3′-nrtA gene was used as a probe. The DIG-DNA labeling and detection kit (Roche, Diagnostics, Mannheim, Germany) was used for signal detection.
Measurement of KA production
The culture supernatant was used for the measurement of KA production. The concentration of KA was determined using the colorimetric method.Citation20) KA forms a chelated compound with ferric ions, thus generating a red color with a peak at approximately 495 nm. Using a standard chemical compound purchased from Wako (Osaka, Japan), the intensity of the peak was proportional to the absorbance in the range from 0.1 to 1.0, which enabled the measurement of KA concentration.
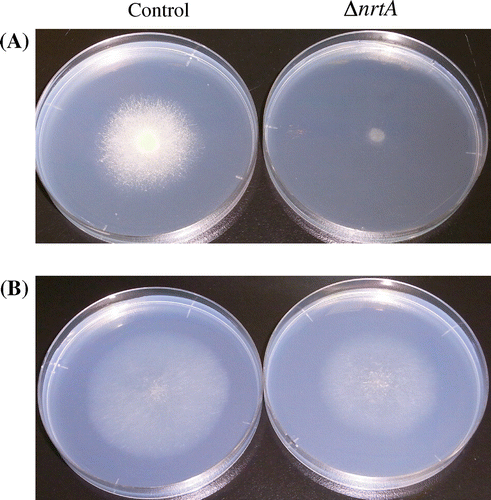
Growth phenotype of the nrtA disruptant
Control and ntrA-disrupted strains were grown at 30 °C for 5 days on minimal medium containing 100 mM NaNO3 or NaNO2
Results and discussion
Analysis of the expression of the nrtA gene
The nrtA (XP_001727608) gene of A. oryzae is homologous to nrtA in other organisms, showing 72, 79, and 99% amino acid identity to nrtA of A. nidulans (XP_658612), Aspergillus fumigatus (XP_752655), and A. flavus (XP_002375899), respectively. NrtA of A. nidulans possesses 12 transmembrane (12-TM) domains.Citation21) Analysis using TMHMM software indicated that the 12-TM domain of NrtA is conserved in A. oryzae. The high degree of identity indicated that nrtA is conserved in Aspergillus species.
In A. nidulans, two nitrate transporters, namely NrtA and NrtB, have been previously identified,Citation11) and these transporters are also present in A. fumigatus. However, A. oryzae and A. flavus possess a single nitrate transporter (NrtA but not NrtB). The nrtA gene of A. oryzae shared 63% identity with nrtB from A. nidulans, indicating that the former strain possesses only a single nitrate transporter gene.
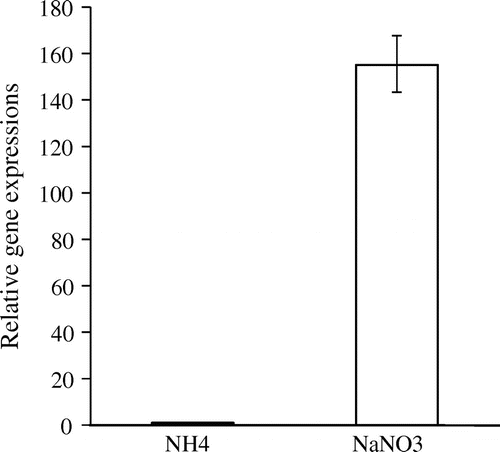
To analyze nrtA expression using real-time PCR, we isolated total RNA and purified mRNA from minimal medium containing 100 mM NaNO3 or 100 mM (NH4)2SO4. The nrtA gene was found to be strongly expressed following the addition of 100 mM NaNO3; the expression of this gene was 155-fold higher than that observed following the addition of 100 mM (NH4)2SO4 (Fig. ). The nonspecific bands obtained using PCR were identified by polyacrylamide gel electrophoresis (data not shown). It is well established that high affinity for NO3− uptake systems in higher plants are rapidly induced by the presence of external NO3−.Citation9) However, nrtA expression was not induced by NaNO2 (data not shown). Therefore, the present real-time PCR data indicate that the nrtA gene of A. oryzae is inducible by NaNO3.
Gene disruption
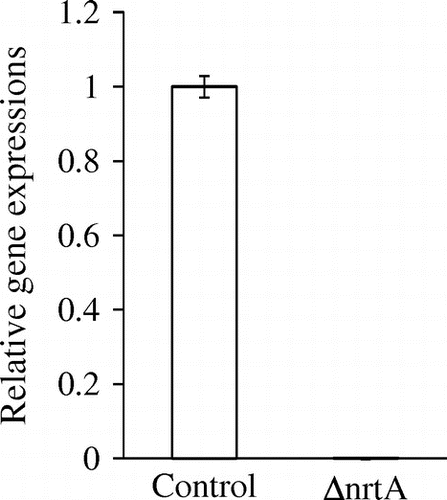
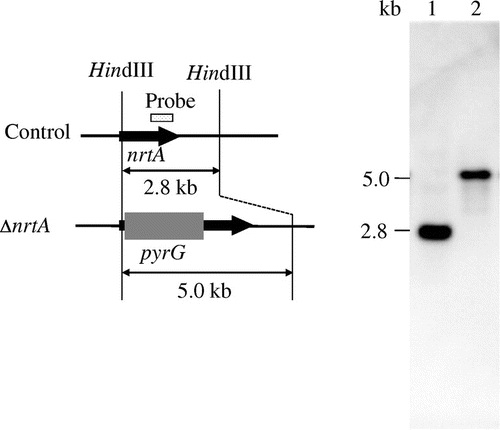
To analyze the function of nrtA in A. oryzae, an nrtA gene disruption cassette was generated by fusion PCR and an nrtA-disrupted mutant of A. oryzae was constructed. Disruption of nrtA was confirmed by PCR analyses (data not shown) and Southern blotting (Fig. ). Southern blot analysis was also used to confirm transformation of the nrtA gene cassette disrupted by homologous recombination. We observed the expected hybridization signals at 2.8 kb and 5.0 kb for digested genomic DNA isolated from the control and disrupted strains, respectively. These results indicated successful homologous recombination at the resident nrtA locus. Furthermore, real-time PCR data showed that nrtA expression in the nrtA-disrupted strain was approximately 833-fold lower than that in the control strain (Fig. ). nrtA expression in the nrtA-disrupted strain was detectable by real-time PCR but was attributed to the amplification of nonspecific bands.
Fig. 2. Southern hybridization analysis of the nrtA-disrupted strain.
Phenotype analysis of the nrtA-disrupted strain
We found that the nrtA-disrupted A. oryzae strain was able to utilize NaNO3 as a nitrogen source. However, this strain showed an almost complete inhibition of growth on NaNO3 plates (Fig. (A)). These data suggest that nrtA is essential for NO3− uptake in A. oryzae. However, two nitrate transporters (NrtA and NrtB) were identified in A. nidulans. nrtA and nrtB single-deletion mutants have been shown to grow on nitrate, whereas the nrtA nrtB double-deletion mutant is unable to grow on nitrate.Citation11) In A. oryzae, only the nrtA-disrupted strain was unable to grow normally when nitrate was the sole nitrogen source. However, the nrtA-disrupted strain exhibited growth on NaNO2 plates (Fig. (B)). These results indicate that the nrtA gene, which encodes a nitrate transporter, is essential for nitrate uptake in A. oryzae, whereas nitrite uptake is mediated by other transporters in this strain.
Fig. 4. Growth phenotype of the nrtA-disrupted strain.
KA production by the nrtA-disrupted strain in the presence of nitrate
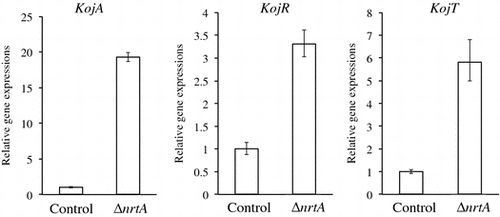
We elucidated the effects of nrtA deletion on KA production in the presence of nitrate. In A. oryzae RIB40, KA production was inhibited by the addition of a small amount of sodium nitrate to the standard medium.Citation6) In the presence of 0.3% sodium nitrate, KA production in the nrtA-deletion strain was detected (17 mM/dry cell weight) 3 days after inoculation, whereas the control strain was found to exhibit KA production (4.6 mM/dry cell weight) (Fig. ). The nrtA-disrupted strain was cultured over 4 days, and the culture supernatant became cloudy. It was very difficult to remove the cloudy supernatant by centrifugation or filtration. Therefore, the supernatant could not be clarified, and we evaluated KA production at 3 days. In the presence of nitrate, KA production by the nrtA-disrupted strain was approximately 3.7-fold higher than that in the control strain. Furthermore, the expression of the KA biosynthesis genes (kojA, kojR, and kojT) increased markedly in the nrtA-disrupted strain in the presence of nitrate (Fig. ). Therefore, in the nrtA-disrupted strain, KA production was not inhibited by the addition of 0.3% sodium nitrate. Moreover, this strain was found to exhibit KA production in the presence of 1.2% sodium nitrate (data not shown). These results demonstrate that the nrtA-disrupted strain is deficient in the uptake of nitrate. As a result, KA production is not significantly affected by the presence of this nitrogen source.
Fig. 5. Effects of nrtA disruption on kojic acid biosynthesis in A. oryzae.
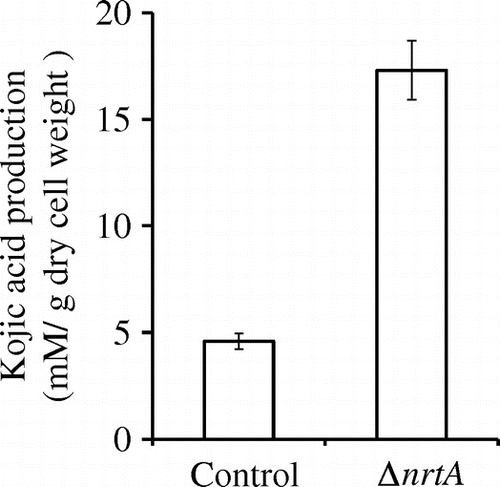
Fig. 6. Evaluation of expression of KA biosynthesis genes (kojA, kojR, and kojT) by real-time PCR.
In the present study, we analyzed the function of the nitrate transporter (NrtA) in A. oryzae. Furthermore, we constructed an nrtA-disrupted strain and evaluated KA production by this strain in the presence of nitrate. These findings are expected to contribute to the optimization of the biosynthesis of kojic acid, which is a commonly used ingredient in skin-lightening cosmetics.
Disclosure statement
No potential conflict of interest was reported by the author.
Funding
This work was supported by the Japan Society for the Promotion of Science (JPSP) KAKENHI [grant number 26450506].
Acknowledgements
We thank Harue Kitagawa and Sanae Tsuji for their technical assistance.
References
- Kang SJ, Choi BR, Lee EK, et al. Inhibitory effect of dried pomegranate concentration powder on melanogenesis in B16F10 melanoma cells; involvement of p38 and PKA signaling pathways. Int. J. Mol. Sci. 2015;16:24219–24242.10.3390/ijms161024219
- Seo SY, Sharma VK, Sharma N. Mushroom tyrosinase: recent prospects. J. Agric. Food Chem. 2003;51:2837–2853.10.1021/jf020826f
- Bentley R. From miso, sake and shoyu to cosmetics: a century of science for kojic acid. Nat. Prod. Rep. 2006;23:1046–1062.10.1039/b603758p
- Rosfarizan M, Ariff AB, Hassan MA, et al. Kojic acid production by Aspergillus flavus using gelatinized and hydrolyzed sago starch as carbon sources. Folia. Microbiol. (Praha). 1998;43:459–464.10.1007/BF02820791
- Yan S, Tang H, Wang S, et al. Improvement of kojic acid production in Aspergillus oryzae B008 mutant strain and its uses in fermentation of concentrated corn stalk hydrolysate. Bioprocess Biosyst. Eng. 2014;37:1095–1103.10.1007/s00449-013-1081-5
- Terabayashi Y, Sano M, Yamane N, et al. Identification and characterization of genes responsible for biosynthesis of kojic acid, an industrially important compound from Aspergillus oryzae. Fungal. Genet. Biol. 2010;47:953–961.10.1016/j.fgb.2010.08.014
- Marui J, Yamane N, Ohashi-Kunihiro S, et al. Kojic acid biosynthesis in Aspergillus oryzae is regulated by a Zn(II)(2)Cys(6) transcriptional activator and induced by kojic acid at the transcriptional level. J. Biosci. Bioeng. 2011;112:40–43.10.1016/j.jbiosc.2011.03.010
- Pao SS, Paulsen IT, Saier MH Jr. Major facilitator superfamily. Microbiol. Mol. Biol. Rev. 1998;62:1–34.
- Forde BG. Nitrate transporters in plants: structure, function and regulation. Biochim. Biophys. Acta. 2000;1465:219–235.10.1016/S0005-2736(00)00140-1
- Unkles SE, Hawker KL, Grieve C, et al. crnA encodes a nitrate transporter in Aspergillus nidulans. Proc. Natl. Acad. Sci. USA. 1991;88:204–208.10.1073/pnas.88.1.204
- Unkles SE, Zhou D, Siddiqi MY, et al. Apparent genetic redundancy facilitates ecological plasticity for nitrate transport. EMBO J. 2001;20:6246–6255.10.1093/emboj/20.22.6246
- Kinghorn JR, Sloan J, Kana’n GJ, et al. Missense mutations that inactivate the Aspergillus nidulans nrtA gene encoding a high-affinity nitrate transporter. Genetics. 2005;169:1369–1377.
- Wang Y, Li W, Siddiqi Y, et al. Nitrite transport is mediated by the nitrite-specific high-affinity NitA transporter and by nitrate transporters NrtA, NrtB in Aspergillus nidulans. Fungal. Genet. Biol. 2008;45:94–102.10.1016/j.fgb.2007.10.001
- Mizutani O, Kudo Y, Saito A, et al. A defect of LigD (human Lig4 homolog) for nonhomologous end joining significantly improves efficiency of gene-targeting in Aspergillus oryzae. Fungal. Genet. Biol. 2008;45:878–889.10.1016/j.fgb.2007.12.010
- Futamura T, Okabe M, Tamura T, et al. Improvement of production of Kojic acid by a mutant strain Aspergillus oryzae, MK107-39. J. Biosci. Bioeng. 2001;91:272–276.10.1016/S1389-1723(01)80133-X
- Wan HM, Chen CC, Giridhar R, et al. Repeated-batch production of kojic acid in a cell-retention fermenter using Aspergillus oryzae M3B9. J. Ind. Microbiol. Biotechnol. 2005;32:227–233.10.1007/s10295-005-0230-5
- Kobayashi A, Sano M, Oda K, et al. The glucoamylase-encoding gene (glaB) is expressed in solid-state culture with a low water content. Biosci. Biotechnol. Biochem. 2007;71:1797–1799.10.1271/bbb.70132
- Tamano K, Satoh Y, Ishii T, et al. The beta-1,3-exoglucanase gene exgA (exg1) of Aspergillus oryzae is required to catabolize extracellular glucan, and is induced in growth on a solid surface. Biosci. Biotechnol. Biochem. 2007;71:926–934.10.1271/bbb.60591
- Takahashi T, Hatamoto O, Koyama Y, et al. Efficient gene disruption in the koji-mold Aspergillus sojae using a novel variation of the positive-negative method. Mol. Genet. Genomics. 2004;272:344–352.10.1007/s00438-004-1062-0
- Bentley R. Preparation and analysis of kojic acid. Methods. Enzymol. 1957;3:238–241.10.1016/S0076-6879(57)03381-9
- Trueman LJ, Richardson A, Forde BG. Molecular cloning of higher plant homologues of the high-affinity nitrate transporters of Chlamydomonas reinhardtii and Aspergillus nidulans. Gene. 1996;175:223–231.10.1016/0378-1119(96)00154-0