Abstract
Aspergillus species are among the most important filamentous fungi from the viewpoints of industry, pathogenesis, and mycotoxin production. Fungal cells are exposed to a variety of environmental stimuli, including changes in osmolality, temperature, and pH, which create stresses that primarily act on fungal cell walls. In addition, fungal cell walls are the first interactions with host cells in either human or plants. Thus, understanding cell wall structure and the mechanism of their biogenesis is important for the industrial, medical, and agricultural fields. Here, we provide a systematic review of fungal cell wall structure and recent findings regarding the cell wall integrity signaling pathways in aspergilli. This accumulated knowledge will be useful for understanding and improving the use of industrial aspergilli fermentation processes as well as treatments for some fungal infections.
Graphical abstract
Fungal cell wall is mainly composed of polysaccharides and essential for cell viability. This review describes the current understanding of cell wall biogenesis in aspergilli.
Fungi are found ubiquitously throughout the world and play an important role in ecosystems as decomposers of plant biomass. In particular, filamentous fungi excel in the ability to secrete a wide variety of hydrolytic enzymes, thereby contributing greatly to terrestrial carbon and nitrogen circulation. Thus, filamentous fungi are indispensable to the maintenance of ecosystems and material circulation on our planet. Nevertheless, they include many phytopathogens that inflict damage on crops. Certain pathogenic species can also infect humans and cause fatal conditions. The risks posed by filamentous fungi are rising in particular as a result of the increasing number of elderly people, HIV sufferers, and others with immunodeficiencies. Whether filamentous fungi bring benefits or harm to our everyday lives depends on human needs and actions, but these fungi produce many different kinds of enzymes, organic acids, and other chemical substances in their own living environments, including many substances of value as raw materials for food processing and pharmaceuticals. A great many fermented foods actually make use of these characteristics of filamentous fungi. The Aspergillus genus includes the koji mold Aspergillus oryzae, A. niger, and other species used in fermentation industries; human pathogens such as A. fumigatus; aflatoxin-producing species such as A. flavus and A. parasiticus; model filamentous fungus A. nidulans, and many other species that exhibit various characteristics. Owing to their importance in industry, medicine, and scientific research, Aspergillus species were among the first filamentous fungi to have had their genomes sequenced and compared.Citation1–4) The sequencing of Aspergillus genomes was a key factor spurring the advances that have been made in research on the cell biology of filamentous fungi over the past decade.
Aspergillus species such as A. oryzae, A. sojae, and A. luchuensis are commonly known in Japan as koji molds for the role they have long played in manufacturing traditional Japanese fermented products such as saké (rice wine), shoyu (soy sauce), miso (soybean paste), and shochu (distilled beverage).Citation5,6) A. oryzae is renowned for its outstanding ability to produce industrially important enzymes such as amylases and proteases.Citation5,7) Koji molds are also noted for their ability to secrete enzymes in higher quantities in solid-state rather than liquid cultures.Citation8) Their long history of utilization in the food industry has, moreover, earned them recognition from the US government’s Food and Drug Administration (FDA) as Generally Recognized as Safe (GRAS) organismsCitation9), and from the World Health Organization (WHO) for their safety.Citation10) The outstanding safety and productivity displayed by A. oryzae makes it an ideal host organism for the production not only of fermented foods, but also of various enzymes and chemicals with pharmacological effects that could be applied to medical treatments in the future.Citation5,7,11,12) In general, the cells of such filamentous fungi need to adapt to changes in temperature and pH, oxidative and osmotic stress, and other changes in the culture environment so as to be able to produce these enzymes and compounds. Such stresses first affect the surface of the fungi. Fungal cells have a cell wall on their surface; for this reason, it is effectively the cell wall that senses changes in the extracellular environment. The stresses detected by the cell wall are transmitted to the cytoplasm and elicit an appropriate cellular response. Also, because filamentous fungi develop filamentous networks of cells known as mycelia as they grow, the cell walls are being constantly synthesized and regenerated during the growth process. As such, the mechanisms regulating cell wall biogenesis, including cell wall stress responses, are the most important and sophisticated mechanisms that affect the survival of filamentous fungi.Citation13–18)
In addition to species that are useful to industry, the Aspergillus genus includes species that produce aflatoxins, which are among the most carcinogenic substances in the natural world, and aflatrem, a tremorgenic mycotoxin; still other Aspergillus species infect humans, often with dire consequences.Citation19,20) A. fumigatus, for example, is the major cause of invasive aspergillosis (IA). It infects and causes severe symptoms in immunocompromised patients, and mortality rate for IA is very high as a result.Citation20) Making contact with host cells is the first decisive step in a pathogen’s attempt to infect a host. The surface structure of A. fumigatus cells, including that of the cell wall surface layer, accordingly plays a critical role in the pathogenic expression of IA. A. fumigatus also produces an extracellular matrix (ECM) composed of polysaccharides, mainly α-1,3-glucan and galactosaminogalactan (GAG). The cell walls of A. fumigatus are coated in this ECM, creating a cell surface structure that is thought to be crucial to pathogenic expression.Citation21)
Cell wall biogenesis and the mechanisms regulating it were first studied in the budding yeast Saccharomyces cerevisiae.Citation22–24) In contrast, our knowledge of filamentous fungi cell wall structure, mechanisms regulating cell wall biogenesis, and cell wall stress responses is limited compared with that of S. cerevisiae. The usefulness and virulence of filamentous fungi as described above are, however, stimulating growing interest in their cell walls. A deeper understanding of the functions of cell walls in filamentous fungi would likely lead not only to the development of new fermented food industry applications, but also to improvements in the microbial production of useful enzymes and other compounds, as well as greater control over pathogenic filamentous fungi. In this article, we will focus on Aspergillus cell wall structure and the mechanisms regulating cell wall biogenesis, introducing recent research findings and reviewing the latest knowledge on the cell walls of filamentous fungi.
I. Cell wall architecture of Aspergillus species
I.i. Overview of cell wall structure
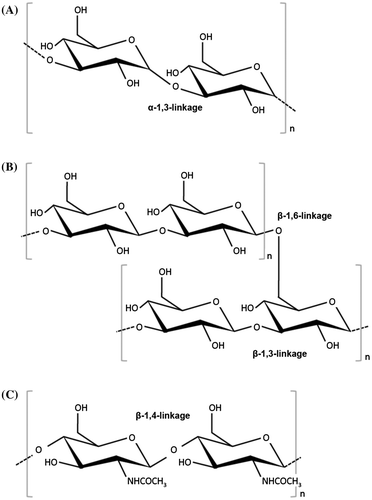
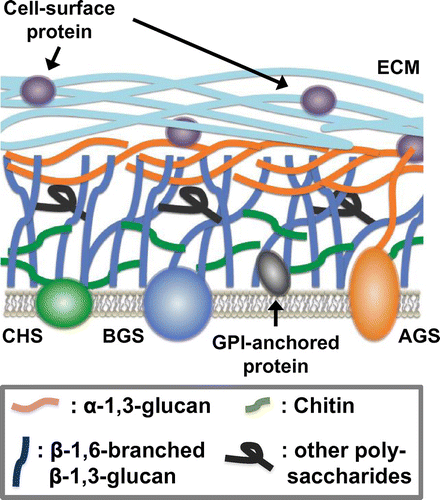
The cell walls of fungi not only play a role in maintaining cell morphology, but also help to protect the cells from various kinds of extracellular environmental stress. As such, cell walls are indispensable to fungi. In their life cycles, fungi are exposed to changes in osmotic pressure, temperature, pH, and various other environmental factors. These environmental changes put stress on the fungi living in the environment concerned, acting first on the cell wall. Fungal cells adapt to the stress of environmental changes by responding in various appropriate ways, including rebuilding and repairing their cell walls. Regarding the mechanisms by which filamentous fungi respond to osmotic and oxidative stress, we refer the reader to the review by Hagiwara et al.Citation25). For both animal and plant pathogens, the cell wall is the first body part to make contact with the host cells. Understanding the structure of the cell wall is accordingly crucial to devise methods of controlling pathogens. The cell walls of filamentous fungi are complex structures composed mainly of polysaccharides (Fig. ). The main polysaccharides involved include α- glucans (mainly α-1,3-glucan (Fig. (A)), but also small quantities of α-1,4-glucan), β- glucans (β-1,3-glucan with β-1,6- branches (Fig. (B)), galactomannan, and chitin (Fig. (C)).Citation26–29) Some fungi also equip themselves with an ECM composed of polysaccharides synthesized de novo in the outer layers of the cell wall (Fig. ).Citation21,30) Galactomannoproteins, GPI-anchored proteins, surface proteins, and other proteins are also found in the cell wall (Fig. ).Citation27–29,31,32) In this section, we review recent findings on the structure, biosynthesis, and biofunctions of the polysaccharides that make up the cell walls of Aspergillus species.
Fig. 1. Schematic illustration of cell wall architecture in Aspergillus species.
I.ii. Cell wall α-1,3-glucan
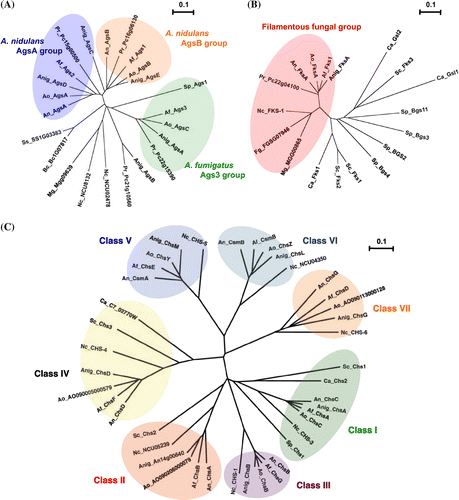
Components of the cell walls of Aspergillus species can be fractionated on the basis of their solubility in alkali.Citation32,33) Alkali-soluble fractions are composed mainly of linear-chain α-1,3-glucan (Fig. (A)). Much of the research conducted so far on the biofunctions of α-1,3-glucan has been carried out on the pathogenic fungi Blastomyces dermatitidis, Cryptococcus neoformans, Histoplasma capsulatumCitation34–36), and the important rice pathogen Magnaporthe griseaCitation37), rather than on Aspergillus spp. This research has, moreover, focused largely on functions related to the pathogenic expression of these species. Research on the functions of α-1,3-glucan in Aspergillus spp. initially focused on analysis of the functions of α-1,3-glucan synthase (AGS) genes.Citation33,38–43) A. fumigatus has three AGS genes, namely ags1, ags2, and ags3 (Fig. (A)).Citation38,40) In the A. fumigatus ags1 (orthologous with A. nidulans agsB) disruptant, the cell wall α-1,3-glucan content decreased by 50%, but no such change in the cell wall α-1,3-glucan content occurred in the A. fumigatus ags2 (orthologous with A. nidulans agsA) disruptant.Citation38) Furthermore, neither disruptant showed any abnormal hyphal apices or abnormal conidiation.Citation38) The remaining A. fumigatus AGS gene, ags3, is a type with no A. nidulans ortholog (Fig. (A)). The ags3 disruptant showed a rise in the expression of ags1, likely owing to overexpression to compensate for the ags3 deficiency.Citation40) This kind of complementary regulation between AGS genes presumably results in maintaining cell wall integrity. The ags1 and ags2 disruptants showed no changes in virulence, but the ags3 disruptant displayed an increase in virulence.Citation40) This change in virulence is probably due to an excessive increase in melanin content in the conidia of the ags3 disruptant, rather than a change in cell wall composition.Citation40) In recent years, multiple disruptants of ags1, ags2, and ags3 have been constructed. A. fumigatus triple AGS disruptants showed no noticeable growth abnormalities when plated, but their cell walls were devoid of α-1,3-glucan.Citation41) They also showed the same kind of decrease in conidiation shown by single ags1 or ags2 disruptants.Citation41) The mycelia of triple AGS disruptants moreover showed increases in β-1,3-glucan and chitin complementary to the disappearance of α-1,3-glucan.Citation41) Whereas germinating conidia of A. Fumigatus wild-type aggregate, those of the triple AGS disruptants did not.Citation41) Evaluation of the virulence of triple AGS disruptants using an experimental murine aspergillosis model showed them to be less virulent than their parental strains.Citation42) A. niger has five AGS genes, namely agsA, agsB, agsC, agsD, and agsE (Fig. (A)).Citation39) Expression of A. niger agsA (orthologous to A. fumigatus ags3) and agsE (orthologous to A. fumigatus ags1, and A. nidulans agsB) is induced in the presence of cell wall stress-inducing compounds such as calcofluor white (CFW), sodium dodecyl sulfate (SDS), and caspofungin, an inhibitor for β-1,3-glucan synthase.Citation39) Several strains of A. nidulans with mutations in the AGS genes agsA and agsB have been created.Citation33,43) The A. nidulans agsA disruptant has so far shown no conspicuous phenotypical differences with the wild type.Citation33) However, agsB disruptants display high sensitivity to congo red (CR) and some types of cell wall degrading enzymes.Citation33) Furthermore, hyphal cells of agsB disruptants dispersed evenly in liquid medium.Citation33) The double agsA/agsB disruptant was also phenotypically identical to the agsB disruptant. Biochemical analysis of the cell wall polysaccharides of these gene disruptants has shown that agsB disruption causes almost total loss of cell wall α-1,3-glucan.Citation33) This α-1,3-glucan is believed to have the same linear chain structure as mutan (which is found in biofilm produced by tooth decay bacterium Streptococcus mutans), because structural analysis showed its structure to match that of mutan enzymatically synthesized using glucanocyl transferase.Citation33) Recently, He et al. have reported that the A. nidulans agsA gene is expressed mainly during conidiation, with two amylase-like proteins (AmyD and AmyG) performing different roles in α-1,3-glucan synthesis.Citation43) Genes encoding AmyD and AmyG form a gene cluster with the gene encoding AgsB, with a gene sequence that is common to some Aspergillus spp, including A. oryzae and A. niger. These two amylase-like proteins are thought to play a role in α-1,3-glucan synthesis, with AmyG being more strongly involved than AmyD.Citation43) In liquid medium, the amyD disruptant shows the same growth properties as the wild type, whereas the amyG disruptant forms the same small pellets of hyphae as the agsB disruptant. Taken together, these findings suggest that α-1,3-glucan performs functions related to pathogenic expression as well as other functions in Aspergillus spp., and that in addition to protecting cells from certain types of cell wall stress, it also helps to promote normal growth and regulate conidiation.
Fig. 3. Phylogenetic tree of α-1,3-glucan synthases (A), β-1,3-glucan synthases (B), and chitin synthases (C) in yeast and filamentous fungi.
I.iii. Cell wall β-glucan and chitin
Different from the alkali-soluble fraction, the alkali-insoluble fraction is composed mainly of β-1,3-glucan, chitin, and galactomannan.Citation32,33) For most fungi, the cell wall central core is composed of a branched β-1,3-/1,6-glucan (Fig. (B)), which is crosslinked to chitin (Fig. (C)) and galactomannan.Citation27–29) Because these alkali-insoluble fractions are thought to be responsible for fungal cell wall rigidity, the biogenesis of β-1,3-glucan and chitin in A. fumigatus has been analyzed in detail.Citation26) The fibrillar core of A. fumigatus cell walls are composed of β-1,3-glucan with ~4% of β-1,6-branch points to which chitin/chitosan, β-1,3-/β-1,4-glucan, and β-1,5-galacto-α-1,2-/α-1,6-mannan (GM) are covalently bound.Citation26–29) In A. fumigatus, β-1,3-glucan is synthesized by a plasma membrane-bound glucan synthase complex, which uses uridine diphosphate (UDP)-glucose as a donor-substrate and extrudes β-1,3-glucan chains through the membrane into the periplasmic space.Citation44,45) A gene homologous to the FKS genes of S. cerevisiae (Fig. (B)), which encodes the putative catalytic subunit of β-1,3-glucan synthase, has been identified in A. fumigatus (Fig. (B)).Citation45) In contrast to yeast, only one FKS gene has been found in the A. fumigatus genome, and this gene is thought to be essential for growth.Citation46) FKS protein of A. fumigatus is an integral membrane protein with putative 16 transmembrane domains, and the A. fumigatus fks1 gene is highly similar to FKS genes of other fungal species (e.g. 90% amino acid identity to A. nidulans FksA) (Fig. (B)).Citation47) As in the case of S. cerevisiae, β-1,3-glucan synthase is regulated by Rho GTPases, and the rho1 gene of A. fumigatus is highly homologous to rho1 of Schizosaccharomyces pombe and S. cerevisiae. Thus, cell wall β-1,3-glucan is biosynthesized by common mechanisms in other fungal species. After linear β-1,3-glucan synthesis, the polysaccharides are remodeled through the combined actions of a specific hydrolase and glucosyltransferase. Subsequently, the modified glucan chains become crosslinked to different polymers, such as chitin and galactomannan, which leads to the complex 3D network of polysaccharides typical in fungal cell walls. However, no enzymes catalyzing such crosslinking activity have been identified in aspergilli.Citation29)
Chitin is a β-1,4-linked homopolymer of N-acetylglucosamine (GlcNAc) whose synthesis is important for hyphal development (Fig. (C)). Chitin synthase (CHS) is an integral plasma membrane protein that catalyzes GlcNAc polymerization from UDP-GlcNAc.Citation48) To date, fungal CHS has been classified into seven classes according to their structural characteristics (Fig. (C)). In this review, the classification proposed by Chigira et al. and Choquer et al. has been accepted.Citation49,50) Class I, II, and IV genes are present in all fungi, whereas classes III, V, VI, and VII are specific to filamentous fungi and certain dimorphic yeasts.Citation32,48,51) Among aspergilli, CHS genes have been comparatively characterized in A. nidulans and A. fumigatus.Citation48) Both these species contain two class III genes.Citation48,52) A. nidulans protein ChsB appears to play important roles in chitin synthesis in hyphal tips and conidia, as well as in polarized hyphal growth.Citation53–55) Deletion of the A. fumigatus class III chsG gene results in similar but less marked defective phenotype, suggesting that its function might be related to those proposed in A. nidulans.Citation56,57) Class V and VI CHSs, CsmA and CsmB, respectively, consist of a C-terminal chitin synthase domain and an N-terminal myosin motor-like domain.Citation58) Myosins are motor proteins that move along actin filaments. Both csmA- and csmB-deletion mutations cause similar growth defects, such as the formation of balloons and intrahyphal hyphae and in hyphal lysis especially under low osmotic conditions, and these mutations are synthetically lethal.Citation58,59) Mutants of the A. fumigatus chsE gene (encoding class V CHS) also show reduced hyphal growth and periodic swellings along hyphal lengths.Citation60) The orthologous genes encoding classes III, V, and VI CHSs have been isolated from other filamentous fungal species and their functions have been investigated.Citation60–70) Results from these observations indicate that CHSs belonging to classes III, V, and VI play crucial roles in hyphal tip growth and maintenance of cell wall integrity. CHSs belonging to classes I, II, and IV tend to make no or relatively small contributions toward chitin biosynthesis in filamentous fungi.Citation48) In addition, the genomes of the yeasts S. cerevisiae, S. pombe, and a dimorphic yeast Candida albicans do not possess CHSs in classes III, V, and VI.Citation48,51) These observations suggest that the CHSs of these three classes likely play critical roles in polarized hyphal growth, especially in filamentous fungi. Taken together, β-1,3-glucan and chitin biosynthesis are fungal specific, and thus, the enzymes related to biosynthesis of these polymers are potential drug targets for therapeutic intervention in fungal diseases affecting humans.
I.iv. Other polysaccharides and proteins
Recent studies have suggested that biofilm formation by A. fumigatus might be one of the most important virulence factors in IA and aspergilloma.Citation21,71) In chronic aspergillosis infections, A. fumigatus develops a biofilm characterized by mycelia embedded in an ECM, referred to as a mycetoma. In both chronic and invasive aspergillosis, hyphae produce an ECM, which is mainly composed of polysaccharides, including GM, GAG, and α-1,3-glucan. It is known that the same polysaccharides are present in ECM found in vivo in chronic and invasive aspergillosis.Citation21) GAG and α-1,3-glucan play major roles in interhyphal adhesion.Citation21,33,41,72) Gravelat et al. have reported that the gene uge3, encoding a fungal epimerase, is required for adherence by mediating the synthesis of GAG, which functions as the dominant adhesive on A. fumigatus and mediates adherence to plastic, fibronectin, and epithelial cells.Citation72) In addition, GAG suppresses host inflammatory responses in vitro and in vivo, in part through masking cell wall β-glucans from recognition by dectin-1. In addition, GAG is essential for full virulence in two murine models of invasive aspergillosis. These findings indicate that GAG plays a pivotal role as a bifunctional virulence factor in the pathogenesis of invasive aspergillosis.Citation72) More recently, a coregulated five-gene cluster has been identified and proposed to encode proteins required for GAG biosynthesis.Citation73) One of these genes, sph3, has been predicted to encode a protein belonging to the spherulin 4 family, whose function is unknown. Analysis of an sph3-deficient mutant in A. fumigatus has demonstrated that the gene is necessary for GAG production. In addition, biochemical analysis using recombinant protein has revealed that Sph3 possesses hydrolytic activity against both purified and cell wall-associated GAG and that the protein is a glycoside hydrolase essential for GAG production, thus defining a new glycoside hydrolase family, GH135.Citation73) The putative GAG biosynthetic gene cluster has been found not only in A. fumigatus but also in other Aspergillus species, such as A. nidulans, niger and oryzae (Miyazawa et al. unpublished results). Thus, common mechanisms for GAG biosynthesis and for subsequent ECM formation are likely employed in Aspergillus species. In addition to polysaccharides, ECM also contains hydrophobins and melanin, both of which are known to be involved in virulence. Better understanding of the biological roles of ECM in filamentous fungi requires further studies, which should include comparative functional analysis of ECM among Aspergillus species.
II. Cell wall integrity signaling system of Aspergillus species
II.i Overview of cell wall integrity signaling
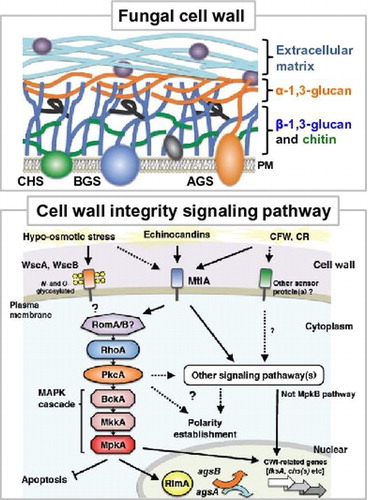
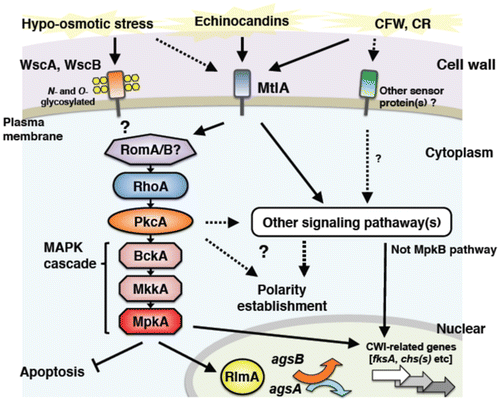
The cell wall integrity (CWI) signaling system that senses cell wall abnormalities and regulates gene responses required to maintain the integrity of cell walls has been studied in detail in the budding yeast S. cerevisiae.Citation22–24,74–87) Cell wall abnormalities are detected by sensor proteins on the membrane. These proteins, which have been labeled WSC after the term “cell wall integrity and stress-response component,” are positioned furthest upstream in the CWI signaling pathway. Sensor proteins Wsc1p, Wsc2p, and Wsc3p are glycosylated transmembrane proteins characterized by their possession of an extracellular cysteine-rich domain, a Ser/Thr-rich region equipped with glycosylation sites, and an intracellular region in the strongly charged C-terminus.Citation23,75–77) Mid2p and Mtl1p, which are also considered to be cell wall stress sensors, are membrane proteins with partially overlapping functions. These are sensors for sensing physical stimulation of the cell wall. They sense morphological changes that accompany growth or pheromone reception as well as cell wall stress caused by exposure to high temperature or high osmotic pressure, and transmit the signals to downstream signaling pathways.Citation75,78,80) The detected cell wall stimuli are transmitted downstream through activation of the CWI signaling pathway. Cell wall stimuli are first transmitted to Rho1p, which is a small G protein.Citation22,80) Rho1p is a low molecular weight GTPase that is activated by the guanosine nucleotide exchange factors (GEFs) Rom1p, Rom2p, and Tus1p.Citation22,81) Its activity is regulated by Bem2p and Sac7p.Citation22,82) Activated Rho1p binds to protein kinase C and activates Pkc1p, thereby inducing activation of the downstream MAP kinase (MAPK) cascade.Citation83) This MAPK cascade is composed of the MAPK kinase kinase Bck1pCitation84), two functionally overlapping MAPK kinases Mkk1p and Mkk2pCitation85), and the MAPK Mpk1p/Slt2p.Citation86) Mpk1p phosphorylates and activates the transcription factor Rlm1p, which regulates the expression of at least 25 cell wall biogenesis-related genes involved in the synthesis of β-1,3-glucan and chitin.Citation87) The genome sequences of Aspergillus spp. have been publishedCitation1–4), and the genes encoding proteins homologous to the constituent factors of the CWI pathway in yeast have been identified in Aspergillus spp. as well.Citation7,15) In this section, we will review the latest information on the mechanisms of CWI pathway regulation in Aspergillus spp.(Fig. ), taking into account the differences in the modes of CWI pathway regulation that have been discovered since genome sequencing.
Fig. 4. Schematic model of cell wall stress signaling in A. nidulans.
II.ii Cell-surface sensors for CWI signaling in Aspergillus species
Progress has been made on analyzing the cell wall stress sensor proteins in Aspergillus species as well.Citation17,18,88–90) Goto et al. have identified the gene that encodes the cell wall stress sensor WscA in A. nidulans (homologous to Wsc1p in S. cerevisiae) (Fig. ).Citation17) A. nidulans WscA contains a Wsc motif rich in cysteine residues, a Ser/Thr-rich region that carries three putative N-glycosylation sites, a transmembrane region, and a cytoplasmic region in the C-terminus. They also discovered that A. nidulans WscA is O-mannosylated in the Ser/Thr-rich region, and that A. nidulans PmtA and PmtC are involved in this reaction.Citation88) PmtA and PmtC are O-mannosyltransferases that are strongly involved in the stability of WscA.Citation91) Glycosylation of proteins also plays an important role in filamentous fungi, as demonstrated by the fact that the pmtA disruptant exhibits abnormal cell morphology and alterations in cell wall composition.Citation92,93) Futagami et al. showed both WscA and WscB sensor proteins to be N- and O-glycosylated and localized in the plasma membrane.Citation17) They also found that wsc gene disruptants (∆wscA and ∆wscB) showed poor growth on a medium, a high frequency of swollen hyphae under hypo-osmotic conditions, and reduced conidiation.Citation17) Interestingly, this abnormal phenotype recovered under osmoregulation with potassium chloride. Furthermore, transcription levels of two α-1,3-glucan synthase genes (agsA and agsB) were altered in the wscA disruptant, resulting in an increase in the amount of alkali-soluble cell wall glucan (i.e. α-1,3-glucan). In contrast, treatment with micafungin, which is a β-1,3-glucan synthase inhibitor, was found to upregulate agsB expression in the wild-type strain and both the ∆wscA and ∆wscB strains. This expression response was dependent on A. nidulans MpkA (orthologous to S. cerevisiae Mpk1p MAPK), indicating that cell wall stress is detected and transmitted to the MpkA pathway even without WscA and WscB.Citation17) A gene encoding a Mid2-like protein with a structure similar to that of the S. cerevisiae cell wall stress sensor Mid2p has also been found in the A. nidulans genome.Citation17,18) Recently, the functions of this A. nidulans Mid2-like protein A (MtlA) were analyzed.Citation89) MtlA protein was found to be involved not only in conidiation, but also in cell wall synthesis and tolerance to stress from CFW, CR, micafungin, and other cell wall synthesis inhibitors. These findings suggest that MtlA protein functions as a cell wall stress sensor in A. nidulans.Citation89)
With regard to the A. fumigatus CWI signaling pathway, Dicht et al. have analyzed the functions of putative cell wall stress sensor proteins Wsc1, Wsc2, Wsc3, and MidA (corresponding to Wsc1p, Wsc2p, Wsc3p, and Mid2p in S. cerevisiae) and Rho GTPases Rho1, Rho2, and Rho4.Citation90) Based on an analysis of the phenotypes of mutants for the genes encoding these CWI sensors and Rho GTPases, they have proposed a model for the A. fumigatus CWI signaling pathway.Citation90) According to the model, cell wall stress caused by CFW, CR, or heat stress is detected by MidA or some other unknown sensor protein. Their signals then probably activate the MpkA pathway through Rho1 and PkcA. Wsc1 functions specifically in response to caspofungin, an echinocandin antifungal agent that inhibits the activity of β-1,3-glucan synthase. A. fumigatus Wsc1, Wsc3, and MidA have overlapping functions, and abnormalities in these proteins affect colony growth and conidiation. Signals from these sensors are transmitted to the MpkA pathway. A. fumigatus Rho2 and Rho4 do not directly affect the activation of MpkA, but are essential to CWI pathway regulation. Rho4 has a role in regulating septum formation, and is also involved in tolerance to β-1,3-glucan synthase inhibitors. These findings suggest that the functions of Wsc and Mid proteins differ between A. fumigatus and A. nidulans.
II.iii Protein kinase C pathway in Aspergillus nidulans
In addition to cell wall stress sensors and Rho GTPases, the functions of protein kinase C (PKC) have been analyzed in several species of filamentous fungi.Citation13,94–98) For example, the PKC of Neurospora crassa, a red bread mold, is essential for growth and reportedly involved in the light response signaling pathway.Citation13,94) In Aspergillus spp., loss of the A. nidulans PKC-encoding gene pkcA (corresponding to pkc1 in yeast) is lethal.Citation96) Yeast in which pkc1 has been deleted is viable in the presence of an osmotic stabilizer, whereas pkcA loss in A. nidulans is lethal regardless of the presence or absence of an osmotic stabilizer.Citation95–98) In contrast, suppression of pkcA gene expression in A. nidulans results in a phenotype showing hypersensitivity to caspofungin, CFW, and other cell wall stress agents, as well as abnormalities in cell wall structure.Citation97,98) These findings suggest that in A. nidulans as well, PkcA is involved in the CWI signaling pathway (Fig. ). Furthermore, A. nidulans PkcA is localized to hyphal apices and growing septa, indicating a role in the formation of septa and phialide apices.Citation97) PkcA is known to have other functions as well, being involved in the regulation of various cellular responses including conidiation and germination, secondary metabolism, and farnesol-induced cell death.Citation15,95–98) Katayama et al. constructed a temperature-sensitive pkcA mutant of A. nidulans that exhibited apoptotic cell death at 42 °C, but showed almost normal growth at 30 °C.Citation99) They further found that PkcA suppresses the induction of apoptosis through the MpkA pathway.Citation99) Additionally, they revealed that although PkcA is also involved in establishing hyphal polarity under conditions of heat stress, MpkA plays no role in this regulation.Citation99) Recently, Katayama et al. constructed a mutant strain of A. nidulans that encodes constitutively activated PkcA.Citation100) Expression of this PkcA-activation mutation in A. nidulans results in increased levels of expression of multiple chitin synthase genes (chsB, chsC, chsD, csmA, and csmB) and the agsB gene.Citation100) This finding indicates that A. nidulans PkcA regulates the transcription of cell wall synthase genes. Whether these transcriptional responses are dependent on the MpkA pathway is a matter that begs further discussion (see the next section), but at least in A. nidulans, PkcA appears to play a central role in the CWI pathway (Fig. ). Moreover, PKC of filamentous fungi is involved in the regulation of multiple cellular responses, and likely plays a key role in cell wall maintenance.
II.iv. MAP kinase pathway for CWI signaling in Aspergillus species
In Aspergillus spp., the genes for encoding MAPK (which corresponds to Mpk1p (Slt2p) in S. cerevisiae) have been isolated.Citation15,101,102) In A. nidulans, analysis of mpkA disruptant phenotypes indicated that this MAP kinase is involved in germination of conidial spores and polarized growth.Citation101) In A. fumigatus, MpkA is reportedly involved in oxidative stress responses, siderophore formation under iron depletion, and regulation of the production of secondary metabolites.Citation102) In addition to these physiological functions of MpkA, the roles played by MpkA in the CWI pathway of Aspergillus spp. have also been analyzed.
Fujioka et al. constructed A. nidulans mpkA, rlmA (orthologous to S. cerevisiae RLM1), and Answi4/Answi6 (orthologous to S. cerevisiae SWI4/SWI6, encoding the Mpk1p-activating TF complex Swi4p-Swi6p in yeast) disruptants that they then used to investigate the cell wall stress response to a β-1,3-glucan synthase inhibitor (micafungin) by analyzing cell wall synthase and mpkA gene transcripts.Citation15) The results showed that the expression levels of almost all cell wall synthase genes were transiently elevated by cell wall stress. This transient elevation in expression was also observed in ∆mpkA and ∆rlmA strains with the exception of two α-1,3-glucan synthase genes (agsA and agsB). This finding suggests that transcription of the β-1,3-glucan synthase gene fksA and several chitin synthase genes (chsA-D, csmA, and csmB) is regulated independently of MpkA-RlmA (Fig. ).Citation15) In A. nidulans, transcription of the agsB gene encoding AgsB, which plays a major role in α-1,3-glucan synthesis, Citation33) is dependent on MpkA regulation.Citation15) The expression level of agsA is extremely low under normal wild-type growth conditions, but was upregulated slightly in ∆mpkA and ∆rlmA strains.Citation15) Fujioka et al. further investigated the regulation mode of the mpkA promoter by constructing an mpkA promoter fused with a GUS reporter. Whereas activity of the GUS reporter system under control of the mpkA promoter was high in the wild-type strain, that of the ∆mpkA strain is significantly low.Citation15) This result indicates that the transcription of mpkA itself is autoregulated via MpkA. Fujioka et al. also confirmed that mpkA expression is independent of RlmA and AnSwi4-AnSwi6.Citation15) Therefore, the expression of the mpkA gene is autoregulated via an unidentified transcription factor regulated by MpkA.Citation15) According to a recent report, MpkA-dependent expression of A. nidulans fksA and csmB was induced by micafungin treatment, and the ∆mpkA strain showed reduced expression of some cell wall genes.Citation89,100) The contradictory nature of these results suggests that there is some room for debate on whether MpkA and RlmA are involved in controlling transcription of fksA and chitin synthase genes in A. nidulans. Recently, Yoshimi et al. discovered that MpkB, a different MAP kinase with the same phosphorylation motif as MpkA, additively contributes to micafungin activity in A. nidulans, but also revealed that MpkB does not directly regulate the CWI pathway.Citation103) This suggests that in A. nidulans, the transcription of chitin synthase and many other cell wall-related genes seems to be regulated not by MpkA or MpkB, but rather by unidentified signaling pathways.
In A. niger, analysis of the transcriptional modes of the genes gfaA and agsA that encode glutamine: fructose-6-phosphate amidotransferase and α-1,3-glucan synthase, respectively, revealed that expression of both is induced in response to cell wall stress.Citation14,104) In silico analysis suggests the presence of RlmA and MsnA (homologous to S. cerevisiae stress response transcription factors Rlm1p and Msn2p/Msn4p) target sites (DNA-binding domains) in the promoter regions of these genes. GUS reporter assays of these promoter regions indicate the presence of one RlmA binding site in the promoter region of A. niger’s agsA gene, indicating that the induction of this gene in response to cell wall stress is fully dependent on this site.Citation14) In the A. niger rlmA deletion strain, agsA induction is completely eliminated and gfaA expression is also reduced under cell wall stress conditions. In the rlmA disruptant, CFW treatment resulted in a rise in cell wall chitin content. The rlmA deletion strain also shows higher sensitivity than wild type toward cell wall stress agents.Citation14) These results indicate that in A. niger, CWI is maintained through RlmA-dependent regulation of the transcription of gfaA, agsA, and other cell wall synthase genes.Citation104) A comparison of the above findings for A. nidulans and A. niger suggests that the two species differ in their MpkA-mediated cell wall stress response mechanisms.
In A. oryzae, the CWI signaling pathway has been studied by analyzing the functions of the subtilisin-like processing protease KexB (orthologous to S. cerevisiae Kex2p)Citation105). S. cerevisiae Kex2p is a Ca2+-dependent transmembrane serine protease that specifically recognizes and cleaves KR and RR sequences of precursor proteins in the Golgi apparatus to produce mature secretory proteins.Citation106,107) α-1,3-glucan, β-1,3-Glucan, chitin, and other polysaccharide synthases as well as Wsc and other sensor proteins are membrane proteins that are probably secreted through exocytosis and function on the membrane. The KexB recognition sequence is actually predicted to be present in the putative amino acid sequence of α-1,3-glucan synthase in A. oryzae (Mizutani et al. unpublished results). These cell wall-related proteins are accordingly thought to mature into normally functioning proteins after undergoing modification by KexB in the Golgi apparatus. The A. oryzae kexB disruptant (ΔkexB) forms shrunken colonies with poor generation of conidia on Czapek Dox (CD) agar plates and hyperbranched mycelia in CD liquid medium.Citation105) The phenotypes of these ΔkexB strains were restored under high osmotic pressure in both solid-state and liquid culture conditions. Comparison of the gene expression profiles of the ΔkexB strain and the wild type with A. oryzae cDNA microarray analysis showed increased transcription of mpkA (the gene that encodes MpkA, a protein thought to be involved in the CWI pathway of koji molds) in the ΔkexB strain compared with the wild-type. Persistent activation of MpkA on CD medium was also evident in the ΔkexB strain. In cells subjected to high osmotic stress, mpkA transcription was downregulated, and even in the ΔkexB strain, sustained phosphorylation of MpkA was not observed. This result does not conflict with the way in which the aforementioned abnormal phenotypes of ∆kexB strain were restored under high osmotic pressure. Higher levels of transcripts for genes encoding β-1,3-glucanosyltransferases, β-1,3-glucan synthase, and chitin synthases were also found in the ΔkexB strain. The above findings suggest that A. oryzae KexB is indispensable to the maintenance of CWI. KexB deletion causes cell wall formation-related maturation abnormalities that are thought to disturb the CWI pathway. It is unclear whether the overexpression of cell wall-related genes observed in the A. oryzae ΔkexB strain is dependent on MpkA. Further research on an A. oryzae ΔkexBΔmpkA strain will be required to clarify this point.
III. Conclusion and prospective
In the past decade, studies of cell wall biogenesis and adaptation mechanisms to cell wall stress in filamentous fungi have advanced greatly because of sequenced genome information and the development of genome-wide analysis tools. In this review, we have described the current understanding of cell wall biogenesis and CWI signaling. Whereas components of CWI signaling in aspergilli are largely similar to those in S. cerevisiae, regulatory targets of CWI in aspergilli seem to differ from those in S. cerevisiae and some cell wall polysaccharides are missing or added among the aspergilli; for instance, α-1,3-glucan and ECM are added in Aspergillus species. Aspergillus fungi evolutionarily must have redesigned their stress response mechanisms in adapting to their harsh environmental niches. As fungal hyphae invade substrates in fermentation, this condition somehow mimics infection process of pathogenic fungi in plants and animals. Therefore, further studies of cell wall biogenesis might mutually accelerate the understanding of adaptation to cell wall stress in fermentation processes as well as infection processes of filamentous fungi.
Disclosure statement
No potential conflict of interest was reported by the authors.
Funding
This work was supported by the Japan Society for the Promotion of Science [KAKENHI Grant-in-Aid for Scientific Research (C) grant number 15K07348].
References
- Machida M, Asai K, Sano M, et al. Genome sequencing and analysis of Aspergillus oryzae. Nature. 2005;438:1157–1161.10.1038/nature04300
- Galagan JE, Calvo SE, Cuomo C, et al. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature. 2005;438:1105–1115.10.1038/nature04341
- Nierman WC, Pain A, Anderson MJ, et al. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature. 2005;438:1151–1156.10.1038/nature04332
- Pel HJ, de Winde JH, Archer DB, et al. Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat. Biotechnol. 2007;25:221–231.10.1038/nbt1282
- Machida M, Yamada O, Gomi K. Genomics of Aspergillus oryzae: learning from the history of Koji mold and exploration of its future. DNA Res. 2008;15:173–183.10.1093/dnares/dsn020
- Hong SB, Lee M, Kim DH, et al. Aspergillus luchuensis, an industrially important black Aspergillus in East Asia. PLoS ONE. 2013;8:e63769.10.1371/journal.pone.0063769
- Kobayashi T, Abe K, Asai K, et al. Genomics of Aspergillus oryzae. Biosci. Biotechnol. Biochem. 2007;71:646–670.10.1271/bbb.60550
- Oda K, Kakizono D, Yamada O, et al. Proteomic analysis of extracellular proteins from Aspergillus oryzae grown under submerged and solid-state culture conditions. Appl. Environ. Microbiol. 2006;72:3448–3457.10.1128/AEM.72.5.3448-3457.2006
- Abe K, Gomi K, Hasegawa F, et al. Impact of Aspergillus oryzae genomics on industrial production of metabolites. Mycopathologia. 2006;162:143–153.10.1007/s11046-006-0049-2
- FAO/WHO. Committee on food additives 31. World Health Organization Technical Report Series: Geneva; 1987
- Sakai K, Kinoshita H, Shimizu T, et al. Construction of a citrinin gene cluster expression system in heterologous Aspergillus oryzae. J. Biosci. Bioeng. 2008;106:466–472.10.1263/jbb.106.466
- Yu J, Proctor R, Brown D, et al. Genomics of economically significant Aspergillus and Fusarium species. In: Arora DK, Khachatourians GG, editors. Applied mycology and biotechnology: fungal genomics. New York, NY: Elsevier; 2003. p. 249–283.
- Arpaia G, Cerri F, Baima S, et al. Involvement of protein kinase C in the response of Neurospora crassa to blue light. Mol. Gen. Genet. 1999;262:314–322.10.1007/s004380051089
- Damveld RA, Arentshorst M, Franken A, et al. The Aspergillus niger MADS-box transcription factor RlmA is required for cell wall reinforcement in response to cell wall stress. Mol. Microbiol. 2005;58:305–319.10.1111/j.1365-2958.2005.04827.x
- Fujioka T, Mizutani O, Furukawa K, et al. MpkA-dependent and -independent cell wall integrity signaling in Aspergillus nidulans. Eukaryot. Cell. 2007;6:1497–1510.10.1128/EC.00281-06
- Colabardini AC, de Castro PA, de Gouvêa PF, et al. Involvement of the Aspergillus nidulans protein kinase C with farnesol tolerance is related to the unfolded protein response. Mol. Microbiol. 2010;78:1259–1279.10.1111/j.1365-2958.2010.07403.x
- Futagami T, Nakao S, Kido Y, et al. Putative stress sensors WscA and WscB are involved in hypo-osmotic and acidic pH stress tolerance in Aspergillus nidulans. Eukaryot. Cell. 2011;10:1504–1515.10.1128/EC.05080-11
- Futagami T, Goto M. Putative cell wall integrity sensor proteins in Aspergillus nidulans. Communicat. Integrat. Biol. 2012;5:206–208.10.4161/cib.18993
- Abe K, Gomi K, Hasegawa F, et al. Impact of Aspergillus oryzae genomics on industrial production of metabolites. Mycopathologia. 2006;162:143–153.10.1007/s11046-006-0049-2
- Latgé JP. Aspergillus fumigatus and Aspergillosis. Clin. Microbiol. Rev. 1999;12:310–350.
- Beauvais A, Fontaine T, Aimanianda V, et al. Aspergillus Cell Wall and Biofilm. Mycopathologia. 2014;178:371–377.10.1007/s11046-014-9766-0
- Levin DE. Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2005;69:262–291.10.1128/MMBR.69.2.262-291.2005
- Levin DE. Regulation of cell wall biogenesis in Saccharomyces cerevisiae: the cell wall integrity signaling pathway. Genetics. 2011;189:1145–1175.10.1534/genetics.111.128264
- Klis FM. Review: cell wall assembly in yeast. Yeast. 1994;10:851–869.10.1002/(ISSN)1097-0061
- Hagiwara D, Yoshimi A, Sakamoto K, et al. Response and adaptation to cell wall stress and osmotic stress in Aspergillus species. In: Takagi H, and Kitagaki H, editors. Stress Biology of Yeasts and Fungi. Tokyo: Springer Japan; 2015. p. 199–218.
- Fontaine T, Simenel C, Dubreucq G, et al. Molecular organization of the alkali-insoluble fraction of Aspergillus fumigatus cell wall. J. Biol. Chem. 2000;275:27594–27607.
- Latgé JP. The cell wall: a carbohydrate armour for the fungal cell. Mol. Microbiol. 2007;66:279–290.10.1111/mmi.2007.66.issue-2
- Latgé JP. Tasting the fungal cell wall. Cellular Microbiol. 2010;12:863–872.10.1111/cmi.2010.12.issue-7
- Gastebois A, Clavaud C, Aimanianda V, et al. Aspergillus fumigatus: cell wall polysaccharides, their biosynthesis and organization. Future Microbiol. 2009;4:583–595.10.2217/fmb.09.29
- Ikeda K, Inoue K, Kitagawa H, et al. The role of the extracellular matrix (ECM) in phytopathogenic fungi: a potential target for disease control. In: Cumagun CJ, editor. Plant Pathology. ISBN: 978-953-51-0489-6, Rijeka: InTech; 2012. p. 131–150.
- Morelle W, Bernard M, Debeaupuis JP, et al. Galactomannoproteins of Aspergillus fumigatus. Eukariot. Cell. 2005;4:1308–1316.
- Bernard M, Latgé JP. Aspergillus fumigatus cell wall: composition and biosynthesis. Med. Mycol. 2001;39:9–17.10.1080/744118873
- Yoshimi A, Sano M, Inaba A, et al. Functional analysis of the α-1,3-glucan synthase genes agsA and agsB in Aspergillus nidulans: AgsB is the major α-1,3-glucan synthase in this fungus. PLoS one. 2013;8:e54893.10.1371/journal.pone.0054893
- Hogan LH, Klein BS. Altered expression of surface alpha-1,3-glucan in genetically related strains of Blastomyces dermatitidis that differ in virulence. Infect Immun. 1994;62:3543–3546.
- Reese AJ, Doering TL. Cell wall α-1,3-glucan is required to anchor the Cryptococcus neoformans capsule. Mol. Microbiol. 2003;50:1401–1409.10.1046/j.1365-2958.2003.03780.x
- Rappleye CA, Goldman WE. Defining virulence genes in the dimorphic fungi. Annu. Rev. Microbiol. 2006;60:281–303.10.1146/annurev.micro.59.030804.121055
- Fujikawa T, Kuga Y, Yano S, et al. Dynamics of cell wall components of Magnaporthe grisea during infectious structure development. Mol. Microbiol. 2009;73:553–570.10.1111/mmi.2009.73.issue-4
- Beauvais A, Maubon D, Park S, et al. Two α(1-3) glucan synthases with different functions in Aspergillus fumigatus. Appl. Environ. Microbiol. 2005;71:1531–1538.10.1128/AEM.71.3.1531-1538.2005
- Damveld RA, vanKuyk PA, Arentshorst M, et al. Expression of agsA, one of five 1,3-α-d-glucan synthase-encoding genes in Aspergillus niger, is induced in response to cell wall stress. Fungal Genet. Biol. 2005;42:165–177.10.1016/j.fgb.2004.11.006
- Maubon D, Park S, Tanguy M, et al. AGS3, an α(1-3)glucan synthase gene family member of Aspergillus fumigatus, modulates mycelium growth in the lung of experimentally infected mice. Fungal Genet. Biol. 2006;43:366–375.10.1016/j.fgb.2006.01.006
- Henry C, Latgé JP, Beauvais A. α1,3 glucans are dispensable in Aspergillus fumigatus. Eukaryot. Cell. 2011;11:26–29.
- Beauvais A, Bozza S, Kniemeyer O, et al. Deletion of the α-(1,3)-glucan synthase genes induces a restructuring of the conidial cell wall responsible for the avirulence of Aspergillus fumigatus. PLOS patho. 2013;9:e1003716.10.1371/journal.ppat.1003716
- He X, Li S, Kaminskyj SGW. Characterization of Aspergillus nidulans α-glucan synthesis: roles for two synthases and two amylases. Mol. Microbiol. 2014;91:579–595.10.1111/mmi.2014.91.issue-3
- Beauvais A, Drake R, Ng K, et al. Characterization of the 1,3-β-glucan synthase of Aspergillus fumigatus. J. Gen. Microbiol. 1993;139:3071–3078.10.1099/00221287-139-12-3071
- Beauvais A, Bruneau JM, Mol PC, et al. Glucan synthase complex of Aspergillus fumigatus. J. Bacteriol. 2000;83:2273–2279.
- Firon A, Beauvais A, Latgé JP, et al. Characterization of essential genes by parasexual genetics in the human fungal pathogen Aspergillus fumigatus. Impact of genomic rearrangements associated with electroporation of DNA. Genetics. 2002;161:1077–1087.
- Douglas CM. Fungal β(1,3)-d-glucan synthesis. Med. Mycol. 2001;39(Suppl 1):55–66.10.1080/mmy.39.1.55.66
- Rogg LE, Fortwendel JR, Juvvadi PR, et al. Regulation of expression, activity and localization of fungal chitin synthases. Med. Mycol. 2012;50:2–17.10.3109/13693786.2011.577104
- Chigira Y, Abe K, Gomi K, et al. chsZ, a gene for a novel class of chitin synthase from Aspergillus oryzae. Curr. Genet. 2002;41:261–267.10.1007/s00294-002-0305-z
- Choquer M, Boccara M, Gonçalves IR, et al. Survey of the Botrytis cinerea chitin synthase multigenic family through the analysis of six euascomycetes genomes. Eur. J. Biochem. 2004;271:2153–2164.10.1111/j.1432-1033.2004.04135.x
- Takeshita N, Wernet V, Tsuizaki M, et al. Transportation of Aspergillus nidulans class III and V chitin synthases to the hyphal tips depends on conventional kinesin. PLoS ONE. 2015;10:e0125937.10.1371/journal.pone.0125937
- Horiuchi H. Functional diversity of chitin synthases of Aspergillus nidulans in hyphal growth, conidiophore development and septum formation. Med. Mycol. 2009;47(s1):S47–S52.10.1080/13693780802213332
- Borgia PT, Iartchouk N, Riggle PJ, et al. The chsB gene of Aspergillus nidulans is necessary for normal hyphal growth and development. Fungal Genet. Biol. 1996;20:193–203.10.1006/fgbi.1996.0035
- Yanai K, Kojima N, Takaya N, et al. Isolation and characterization of two chitin synthase genes from Aspergillus nidulans. Biosci. Biotechnol. Biochem. 1994;58:1828–1835.10.1271/bbb.58.1828
- Fukuda K, Yamada K, Deoka K, et al. Class III chitin synthase ChsB of Aspergillus nidulans localizes at the sites of polarized cell wall synthesis and is required for conidial development. Eukaryot. Cell. 2009;8:945–956.10.1128/EC.00326-08
- Mellado E, Specht CA, Robbins PW, et al. Cloning and characterization of chsD, a chitin synthase-like gene of Aspergillus fumigatus. FEMS Microbiol. Lett. 1996;143:69–76.10.1111/fml.1996.143.issue-1
- Mellado E, Dubreucq G, Mol P, et al. Cell wall biogenesis in a double chitin synthase mutant (chsG-/chsE-) of Aspergillus fumigatus. Fungal Genet. Biol. 2003;38:98–109.10.1016/S1087-1845(02)00516-9
- Takeshita N, Yamashita S, Ohta A, et al. Aspergillus nidulans class V and VI chitin synthases CsmA and CsmB, each with a myosin motor-like domain, perform compensatory functions that are essential for hyphal tip growth. Mol. Mocrobiol. 2006;59:1380–1394.10.1111/j.1365-2958.2006.05030.x
- Horiuchi H, Fujiwara M, Yamashita S, et al. Proliferation of intrahyphal hyphae caused bydisruption of csmA which encodes a class V-chitin synthase with a myosin motor-like domain in Aspergillus nidulans. J Bacteriol. 1999;181:3721–3729.
- Aufauvre-Brown A, Mellado E, Gow NAR, et al. Aspergillus fumigatus chsE: a gene related to CHS3 of Saccharomyces cerevisiae and important for hyphal growth and conidiophore development but not pathogenicity. Fungal Genet. Biol. 1997;21:141–152.10.1006/fgbi.1997.0959
- Kim J-E, Lee H-J, Lee J, et al. Gibberella zeae chitin synthase genes, GzCHS5 and GzCHS7, are required for hyphal growth, perithecia formation, and pathogenicity. Curr. Genet. 2009;55:449–459.10.1007/s00294-009-0258-6
- Kong L-A, Yang J, Li G-T, et al. Different chitin synthase genes are required for various developmental and plant infection processes in the rice blast fungus Magnaporthe oryzae. PLoS Patho. 2012;8:e1002526.10.1371/journal.ppat.1002526
- Larson TM, Kendra DF, Busman M, et al. Fusarium verticillioides chitin synthases CHS5 and CHS7 are required for normal growth and pathogenicity. Curr. Genet. 2011;57:177–189.10.1007/s00294-011-0334-6
- Liu HB, Kauffman S, Becker JM, et al. Wangiella (Exophiala) dermatitidis WdChs5p, a class V chitin synthase, is essential for sustained cell growth at temperature of infection. Eukaryot. Cell. 2011;3:40–51.
- Mellado E, Aufauvre-Brown A, Gow NAR, et al. The Aspergillus fumigatus chsC and chsG genes encode class III chitin synthases with different functions. Mol. Microbiol. 1996;20:667–679.10.1046/j.1365-2958.1996.5571084.x
- Pareja-Jaime Y, Martín-Urdíroz M, Roncero MI, et al. Chitin synthase-deficient mutant of Fusarium oxysporum elicits tomato plant defence response and protects against wild-type infection. Mol. Plant Pathol. 2010;11:479–493.10.1111/j.1364-3703.2010.00624.x
- Treitschke S, Doehlemann G, Schuster M, et al. The myosin motor domain of fungal chitin synthase V is dispensable for vesicle motility but required for virulence of the maize pathogen Ustilago maydis. Plant Cell. 2010;22:2476–2494.10.1105/tpc.110.075028
- Werner S, Sugui JA, Steinberg G, et al. A chitin synthase with a myosin-like motor domain is essential for hyphal growth, appressorium differentiation, and pathogenicity of the maize anthracnose fungus Colletotrichum graminicola. Mol. Plant-Microbe Interact. 2007;20:1555–1567.10.1094/MPMI-20-12-1555
- Soulie MC, Perino C, Piffeteau A, et al. Botrytis cinerea virulence is drastically reduced after disruption of chitin synthase class III gene (Bcchs3a). Cell Microbiol. 2006;8:1310–1321.10.1111/cmi.2006.8.issue-8
- Sheng W, Yamashita S, Ohta A, et al. Functional differentiation of chitin synthases in Yarrowia lipolytica. Biosci. Biotechnol. Biochem. 2013;77:1275–1281.10.1271/bbb.130111
- Kaur S, Singh S. Biofilm formation by Aspergillus fumigatus. Med. Mycol. 2014;52:2–9.
- Gravelat FN, Beauvais A, Liu H, et al. Aspergillus galactosaminogalactan mediates adherence to host constituents and conceals hyphal β-glucan from the immune system. PLoS patho. 2013;9:e1003575.10.1371/journal.ppat.1003575
- Bamford NC, Snarr BD, Gravelat FN, et al. Sph3 is glycoside hydrolase required for biosynthesis of galactosaminogalactan in Aspergillus fumigatus. J Biol. Chem. 2015;290:27438–27450.
- Heinisch JJ, Lorberg A, Schmitz HP, et al. The protein kinase C-mediated MAP kinase pathway involved in the maintenance of cellular integrity in Saccharomyces cerevisiae. Mol. Microbiol. 1999;32:671–680.10.1046/j.1365-2958.1999.01375.x
- Verna J, Lodder A, Lee K, et al. A family of genes required for maintenance of cell wall integrity and for the stress response in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 1997;94:13804–13809.10.1073/pnas.94.25.13804
- Lodder AL, Lee TK, Ballester R. Characterization of the Wsc1 protein, a putative receptor in the stress response of Saccharomyces cerevisiae. Genetics. 1999;152:1487–1499.
- Rodicio R, Heinisch JJ. Together we are strong—cell wall integrity sensors in yeasts. Yeast. 2010;27:531–540.10.1002/yea.1785
- Ketela T, Green R, Bussey H. Saccharomyces cerevisiae Mid2p is a potential cell wall stress sensor and upstream activator of the PKC1-MPK1 cell integrity pathway. J. Bacteriol. 1999;181:3330–3340.
- Rajavel M, Philip B, Buehrer BM, et al. Mid2 is a putative sensor for cell integrity signaling in Saccharomyces cerevisiae. Mol. Cell Biol. 1999;19:3969–3976.10.1128/MCB.19.6.3969
- Levin DE, Fields FO, Kunisawa R, et al. A candidate protein kinase C gene, PKC1, is required for the S. cerevisiae cell cycle. Eukaryot. Cell. 1990;62:213–224.10.1016/0092-8674(90)90360-Q
- Philip B, Levin DE. Wsc1 and Mid2 are cell surface sensors for cell wall integrity signaling that act through Rom2, a guanine nucleotide exchange factor for Rho1. Mol. Cell Biol. 2001;21:271–280.10.1128/MCB.21.1.271-280.2001
- Schmidt A, Bickle M, Beck T, et al. The yeast phosphatidylinositol kinase homolog TOR2 activates RHO1 and RHO2 via the exchange factor ROM2. Cell. 1997;88:531–542.10.1016/S0092-8674(00)81893-0
- Kamada Y, Qadota H, Python CP, et al. Activation of yeast protein kinase C by Rho1 GTPase. J. Biol. Chem. 1996;271:9193–9196.
- Lee KS, Levin DE. Dominant mutations in a gene encoding a putative protein kinase (BCK1) bypass the requirement for a Saccharomyces cerevisiae protein kinase C homolog. Mol. Cell Biol. 1992;12:172–182.10.1128/MCB.12.1.172
- Irie K, Takase M, Lee KS, et al. MKK1 and MKK2, which encode Saccharomyces cerevisiae mitogen-activated protein kinase kinase homologs, function in the pathway mediated by protein kinase C. Mol. Cell Biol. 1993;13:3076–3083.
- Martin H, Arroyo J, Sanchez M, et al. Activity of the yeast MAP kinase homologue Slt2 is critically required for cell integrity at 37 degrees C. Mol. Gen. Genet. 1993;241:177–184.10.1007/BF00280215
- Jung US, Levin DE. Genome-wide analysis of gene expression regulated by the yeast cell wall integrity signalling pathway. Mol. Microbiol. 1999;34:1049–1057.10.1046/j.1365-2958.1999.01667.x
- Goto M, Harada Y, Oka T, et al. Protein O-mannosyltransferases B and C support hyphal development and differentiation in Aspergillus nidulans. Eukaryot. Cell. 2009;8:1465–1474.10.1128/EC.00371-08
- Futagami T, Seto K, Kajiwara Y, et al. The putative stress sensor protein MtlA is required for conidia formation, cell wall stress tolerance, and cell wall integrity in Aspergillus nidulans. Biosci. Biothecnol. Tol. Biochem. 2014;78:326–335.10.1080/09168451.2014.878218
- Dichtl K, Helmschrott C, Dirr F, et al. Deciphering cell wall integrity signalling in Aspergillus fumigatus: identification and functional characterization of cell wall stress sensors and relevant Rho GTPases. Mol. Microbiol. 2012;83:506–519.10.1111/mmi.2012.83.issue-3
- Kriangkripipat T, Momany M. Aspergillus nidulans protein O-mannosyltransferases play roles in cell wall integrity and developmental patterning. Eukaryot. Cell. 2009;8:1475–1485.10.1128/EC.00040-09
- Oka T, Hamaguchi T, Sameshima Y, et al. Molecular characterization of protein O-mannosyltransferase and its involvement in cell-wall synthesis in Aspergillus nidulans. Microbiology. 2004;150:1973–1982.10.1099/mic.0.27005-0
- GOTO M. Protein O-glycosylation in fungi: diverse structures and multiple functions. Biosci. Biotechnol. Biochem. 2007;71:1415–1427.10.1271/bbb.70080
- Franchi L, Fulci V, Macino G. Protein kinase C modulates light responses in Neurospora by regulating the blue light photoreceptor WC-1. Mol. Microbiol. 2005;56:334–345.10.1111/j.1365-2958.2005.04545.x
- Herrmann M, Sprote P, Brakhage AA. Protein kinase C (PkcA) of Aspergillus nidulans is involved in penicillin production. Appl. Environ. Microbiol. 2006;72:2957–2970.10.1128/AEM.72.4.2957-2970.2006
- Ichinomiya M, Uchida H, Koshi Y, et al. A protein kinase C-encoding gene, pkcA , is essential to the viability of the filamentous fungus Aspergillus nidulans. Biosci. Biotechnol. Biochem. 2007;71:2787–2799.10.1271/bbb.70409
- Ronen R, Sharon H, Levdansky E, et al. The Aspergillus nidulans pkcA gene is involved in polarized growth, morphogenesis and maintenance of cell wall integrity. Curr. Genet. 2007;51:321–329.10.1007/s00294-007-0129-y
- Teepe AG, Loprete DM, He Z, et al. The protein kinase C orthologue PkcA plays a role in cell wall integrity and polarized growth in Aspergillus nidulans. Fungal Genet. Biol. 2007;44:554–562.10.1016/j.fgb.2006.10.001
- Katayama T, Uchida H, Ohta A, et al. Involvement of protein kinase C in the suppression of apoptosis and in polarity establishment in Aspergillus nidulans under conditions of heat stress. PLoS ONE. 2012;7:e50503.10.1371/journal.pone.0050503
- Katayama T, Ohta A, Horiuchi H. Protein kinase C regulates the expression of cell wall-related genes in RlmA-dependent and independent manners in Aspergillus nidulans. Biosci. Biothechnol. Biochem. 2015;79:321–330.10.1080/09168451.2014.973365
- Bussink HJ, Osmani SA. A mitogen-activated protein kinase (MPKA) is involved in polarized growth in the filamentous fungus, Aspergillus nidulans. FEMS Microbiol. Lett. 1999;173:117–125.10.1111/fml.1999.173.issue-1
- Jain R, Valiante V, Remme N, et al. The MAP kinase MpkA controls cell wall integrity, oxidative stress response, gliotoxin production and iron adaptation in Aspergillus fumigatus. Mol. Microbiol. 2011;82:39–53.10.1111/mmi.2011.82.issue-1
- Yoshimi A, Fujioka T, Mizutani O, et al. Mitogen-activated protein kinases MpkA and MpkB independently affect micafungin sensitivity in Aspergillus nidulans. Biosci. Biothechnol. Biochem. 2015;79:836–844.10.1080/09168451.2014.998619
- Ram AF, Arentshorst M, Damveld RA, et al. The cell wall stress response in Aspergillus niger involves increased expression of the glutamine : fructose-6-phosphate amidotransferase-encoding gene (gfaA) and increased deposition of chitin in the cell wall. Microbiology. 2004;150:3315–3326.10.1099/mic.0.27249-0
- Mizutani O, Nojima A, Yamamoto M, et al. Disordered cell integrity signaling caused by disruption of the kexB gene in Aspergillus oryzae. Eukaryot. Cell. 2004;3:1036–1048.10.1128/EC.3.4.1036-1048.2004
- Fuller RS, Brake AJ, Thorner J. Yeast prohormone processing enzyme (KEX2 gene product) is a Ca2+-dependent serine protease. Proc. Natl. Acad. Sci. USA. 1989;86:1434–1438.10.1073/pnas.86.5.1434
- Redding K, Holcomb C, Fuller RS. Immunolocalization of Kex2 protease identifies a putative late Golgi compartment in the yeast Saccharomyces cerevisiae. J. Cell Biol. 1991;113:527–538.10.1083/jcb.113.3.527