Abstract
Although Ewing sarcoma protein (EWS) is known to be glycosylated by O-linked β-N-acetylglucosamine (O-GlcNAc), the dynamics and stoichiometry of its glycosylation remain obscure. Here, we report a dynamic change in the glycosylation stoichiometry of EWS species during neuronal differentiation of embryonic carcinoma P19 cells. Our findings suggest that O-GlcNAc glycosylation participates in the regulation of EWS functions in neuronal cells.
The Ewing sarcoma protein (EWS) is one of the three members of the FET (FUS/EWS/TAF15) family of nucleic acid-binding proteins and has been suggested to play multiple roles in transcription, and RNA processing and transport.Citation1) A previous study showed that Ews-deficient mice were born at the expected Mendelian ratio but were smaller than their littermates and had a high rate (about 90%) of postnatal mortality prior to weaning.Citation2) Analyses of these mice showed that Ews is essential for meiosis, pre-B lymphocyte development, prevention of premature cellular senescence in fibroblasts and hematopoietic stem progenitor cells, early brown fat lineage determination, and white adipocyte differentiation.Citation2–5) It has also been reported that knockdown of the two zebrafish EWS orthologs, ewsr1a and ewsr1b, leads to a reduced number of proneural cells in the zebrafish central nervous system during early embryonic development.Citation6) These lines of evidence suggested that EWS plays a key role in the development and differentiation of a variety of cell lineages including neuronal cells. In addition, EWS is implicated in neurodegenerative diseases, such as frontotemporal lobar degeneration.Citation7) Therefore, understanding the regulatory mechanisms of multiple EWS roles in the nervous system is important for developing therapeutics for neurodegenerative diseases. Although EWS has been identified as an O-linked β-N-acetylglucosaminylated (O-GlcNAcylated) proteinCitation8–11) and neuronal differentiation has been found to stimulate EWS glycosylation,Citation8) the glycosylation stoichiometry of EWS species, especially in neuronal cells, has not been examined. The present study was conducted to investigate the extent of glycosylated EWS species during neuronal differentiation.
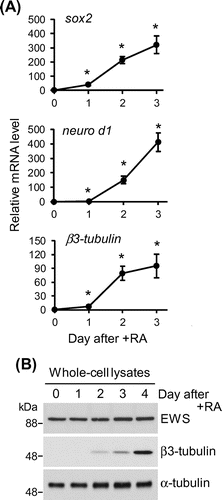
Mouse embryonic carcinoma P19 cells were grown in Dulbecco’s modified Eagle’s medium (Wako Pure Chemicals) supplemented with 10% heat-inactivated fetal bovine serum, MEM non-essential amino acids (Wako Pure Chemicals), 100 U/mL penicillin, and 100 μg/mL streptomycin at 37 °C under humidified air with 5% CO2. To induce neuronal differentiation, 2 × 105 cells/mL of the cells were treated with 5 μM all-trans retinoic acid and plated on bacterial-grade dishes to culture in suspension. The aggregated embryoid body-like cells were again fed with differentiation medium two days later. The expression profiles of neuronal differentiation markers were analyzed at 24-h intervals for 72 h by quantitative RT-PCR. All primers used are shown in Table . Regarding the products of the genes analyzed, Sox2 is an essential transcription factor for the early stages of neuronal differentiation,Citation12) and Neuro D1Citation13) and β3-tubulinCitation14) are the earliest markers of neurons. The expression of these neuronal differentiation markers increased significantly beginning on day one (Fig. (A)). As shown in Fig. (B), the expression levels of EWS did not change before or after the induction of neuronal differentiation and were constant during the early stages of the neuronal differentiation of P19 cells.
Table 1. Primers used in this study.
Fig. 1. The expression profile of EWS during the early stages of neuronal differentiation.
We confirmed the glycosylation of immunoprecipitated EWS from P19 cells by an O-GlcNAc-specific antibody (data not shown) and then examined the glycosylation stoichiometry of EWS during neuronal differentiation using chemoenzymatic labeling of O-GlcNAc residues.Citation15,16) In brief, an engineered mutant, β-1,4-galactosyltransferase (Y289L), catalyzes the transfer of azide-modified galactose (GalNAz) from UDP-GalNAz to terminal GlcNAc residues of glycoproteins. A 5-kDa polyethylene glycol (PEG) alkyne tag is then reacted with the transferred GalNAz through Huisgen azide-alkyne cycloaddition. Since EWS is a nucleocytoplasmic O-GlcNAcylated protein and is not physiologically modified by N-linked glycans which might have terminal GlcNAc residues, this approach enables separation of the stoichiometrically different O-GlcNAcylated species upon SDS–PAGE and immunoblotting with an antibody against EWS and quantification of the O-GlcNAcylation stoichiometry of EWS by densitometry.
To characterize the O-GlcNAcylation stoichiometry of EWS in P19 cells, cell lysates were prepared at 24-h intervals during neuronal differentiation and chemoenzymatically labeled glycoproteins were separated by SDS–PAGE and probed for EWS by immunoblotting. As shown in Fig. (A), two antibody-reactive bands were detected above the unmodified EWS band in undifferentiated cells, although the upper one was faint (lane day 0, + GalNAz-PEG reaction). The molecular weights of these bands were approximately 5 and 10 kDa higher than the unmodified EWS band, corresponding to EWS species O-GlcNAcylated at one and two sites, respectively. Densitometric analysis showed that unmodified, monoglycosylated, and diglycosylated species represented 71.6%, 26.5%, and 1.9% of the total EWS population, respectively (Fig. (B)).
Fig. 2. O-GlcNAcylation stoichiometry of EWS is regulated by neuronal differentiation in P19 cells.
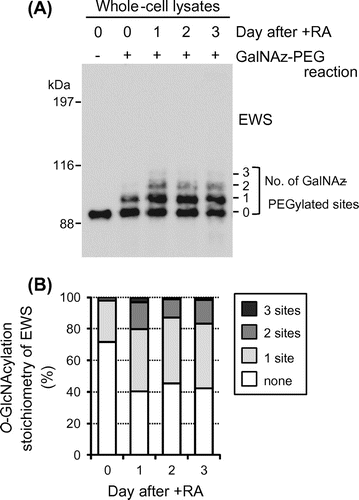
Interestingly, the O-GlcNAcylation stoichiometry of EWS changed greatly upon induction of neuronal differentiation. From days 1 to 3, unmodified EWS, monoglycosylated EWS, and diglycosylated EWS represented 40–45%, 39–42%, and 12–17% of the total EWS population, respectively (Fig. (A) and (B)). In addition, triglycosylated species (EWS species O-GlcNAcylated at three sites) were observed during neuronal differentiation (1–3% of the total EWS population; Fig. (A) and (B)). These results indicate that the majority of EWS species are glycosylated during neuronal differentiation and O-GlcNAcylation is part of the regulatory mechanism of EWS functions in neuronal cells.
Previously, O-GlcNAcylation of the N-terminal EWS transactivation domain of the EWS-FLI1 oncoprotein has been shown to be involved in the transcriptional activity of EWS-FLI1.Citation17) We have reported that in silico O-GlcNAcylation site analysis using the YinOYang 1.2 server (http://www.cbs.dtu.dk/services/YinOYang/)Citation18) predicted that 54 Ser/Thr residues of mouse EWS are the possible glycosylation sites and that these putative sites are unevenly distributed in the N-terminus.Citation11) We therefore speculate that EWS plays a role in transcription in an O-GlcNAcylation-dependent manner in neuronal cells. We are continuing to identify the glycosylation sites of EWS and to investigate the importance of glycosylation for EWS functions in neuronal cells.
Author contributions
Kazuo Kamemura and Hiromi Abe conducted experiments. Kazuo Kamemura wrote the paper.
Disclosure statement
No potential conflict of interest was reported by the authors.
Funding
This work was supported in part by the Japan Society for the Promotion of Science (JSPS) KAKENHI No. [15K00412], [16K07700] to K.K.
Notes
Abbreviations: EWS, Ewing sarcoma protein; GalNAz, azide-modified galactose; O-GlcNAc, O-linked β-N-acetylglucosamine; O-GlcNAcylation, O-GlcNAc glycosylation; PEG, polyethylene glycol; RA, all-trans-retinoic acid.
References
- Schwartz JC, Cech TR, Parker RR. Biochemical properties and biological functions of FET proteins. Annu. Rev. Biochem. 2015;84:3.1–3.25.
- Li H, Watford W, Li C, et al. Ewing sarcoma gene EWS is essential for meiosis and B lymphocyte development. J. Clin. Invest. 2007;117:1314–1323.10.1172/JCI31222
- Cho J, Shen H, Yu H, et al. Ewing sarcoma gene Ews regulates hematopoietic stem cell senescence. Blood. 2011;117:1156–1166.10.1182/blood-2010-04-279349
- Park JH, Kang HJ, Kang SI, et al. A multifunctional protein, EWS, is essential for early brown fat lineage determination. Dev. Cell. 2013;26:393–404.10.1016/j.devcel.2013.07.002
- Park JH, Lee SB. An essential role for Ewing sarcoma gene (EWS) in early white adipogenesis. Obesity. 2015;23:138–144.10.1002/oby.20934
- Azuma M, Embree LJ, Sabaawy H, et al. Ewing sarcoma protein Ewsr1 maintains mitotic integrity and proneural cell survival in the zebrafish embryo. PLoS One. 2007;2:e979.10.1371/journal.pone.0000979
- Dormann D, Haass C. Fused in sarcoma (FUS): an oncogene goes awry in neurodegeneration. Mol. Cell. Neurosci. 2013;56:475–486.10.1016/j.mcn.2013.03.006
- Matsuoka Y, Matsuoka Y, Shibata S, et al. Identification of Ewing's sarcoma gene product as a glycoprotein using a monoclonal antibody that recognizes an immunodeterminant containing O-linked N-acetylglucosamine moiety. Hybrid. Hybrydomics. 2002;21:233–236.10.1089/153685902760213831
- Wells L, Vosseller K, Cole RN, et al. Mapping sites of O-GlcNAc modification using affinity tags for serine and threonine post-translational modifications. Mol. Cell. Proteomics. 2002;1:791–804.10.1074/mcp.M200048-MCP200
- Ishihara K, Takahashi I, Tsuchiya Y, et al. Characteristic increase in nucleocytoplasmic protein glycosylation by O-GlcNAc in 3T3-L1 adipocyte differentiation. Biochem. Biophys. Res. Commun. 2010;398:489–494.10.1016/j.bbrc.2010.06.105
- Li Q, Kamemura K. Adipogenesis stimulates the nuclear localization of EWS with an increase in its O-GlcNAc glycosylation in 3T3-L1 cells. Biochem. Biophys. Res. Commun. 2014;450:588–592.10.1016/j.bbrc.2014.06.013
- Kishi M, Mizuseki K, Sasai N, et al. Requirement of Sox2-mediated signaling for differentiation of early Xenopus neuroectoderm. Development. 2000;127:791–800.
- Przyborski SA, Morton IE, Wood A, et al. Developmental regulation of neurogenesis in the pluripotent human embryonal carcinoma cell line NTERA-2. Eur. J. Neurosci. 2000;12:3521–3528.10.1046/j.1460-9568.2000.00230.x
- Lee MK, Tuttle JB, Rebhun L, et al. The expression and posttranslational modification of a neuron-specific β-tubulin isotype during chick embryogenesis. Cell Motil. Cytoskeleton. 1990;17:118–132.10.1002/(ISSN)1097-0169
- Rexach JE, Rogers CJ, Yu S-H, et al. Quantification of O-glycosylation stoichiometry and dynamics using resolvable mass tags. Nat. Chem. Biol. 2010;6:645–651.10.1038/nchembio.412
- Nagel AK, Ball LE. O-GlcNAc modification of the runt-related transcription factor 2 (Runx2) links osteogenesis and nutrient metabolism in bone marrow mesenchymal stem cells. Mol. Cell. Proteomics. 2014;13:3381–3395.10.1074/mcp.M114.040691
- Bachmaier R, Aryee DNT, Jug G, et al. O-GlcNAcylation is involved in the transcriptional activity of EWS-FLI1 in Ewing’s sarcoma. Oncogene. 2009;28:1280–1284.10.1038/onc.2008.484
- Gupta R, Brunak S. Prediction of glycosylation across the human proteome and the correlation to protein function. Pac. Symp. Biocomput. 2002;7:310–322.
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408.10.1006/meth.2001.1262
