Abstract
Many microorganisms harbor genes necessary to synthesize biodegradable plastics known as polyhydroxyalkanoates (PHAs). We surveyed a genomic database and discovered a new cluster of class IV PHA synthase genes (phaRC). These genes are different in sequence and operon structure from any previously reported PHA synthase. The newly discovered PhaRC synthase was demonstrated to produce PHAs in recombinant Escherichia coli.
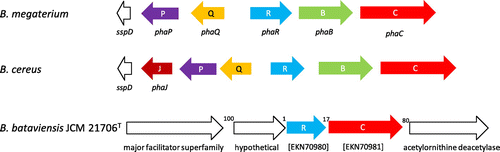
Polyhydroxyalkanoates (PHAs) are biopolyesters produced by many bacteria and some archaea to store carbon and energy.Citation1) They are studied with the focus on applications for the biodegradable plastics industry, particularly for medical applications such as tissue engineering and drug delivery.Citation2) To date, PHA synthases are classified into four groups on the basis of their subunit composition, operon structure, and substrate specificity.Citation3) Class I, III, and IV synthases have substrate specificity toward short-chain-length hydroxyacyl monomer units (C3–C5), whereas class II synthases have substrate specificity toward medium-chain-length hydroxyacyl monomer units (C6–C14).Citation3) Class IV PHA synthases are heterodimeric and have been reported in a wide variety of Bacillus species, which are Gram-positive, rod-shaped bacteria, and considered to be safe PHA production hosts because of the absence of lipopolysaccharide, a pyrogenic endotoxin in humans.Citation4) Recently, our group surveyed a genome database and proposed three subgroups in class IV synthases based on their operon structures (Fig. ), two from B. megaterium and B. cereus have been cloned and studied for cellular and molecular aspects;Citation5) however, the third one has not been previously described and it was unknown as to whether or not it is functional. In this study, we cloned the phaRC synthase genes (PhaC and PhaR are the subunits for a heterodimeric PHA synthase) from B. bataviensis JCM 21706T as a prototype of the new subgroup, heterologously expressed the genes in Escherichia coli JM109, and confirmed the ability of this new PHA synthase to produce PHA polymer.
Fig. 1. Schematic diagram of pha operons on the genome sequences of B. bataviensis JCM 21706T and two representative species.
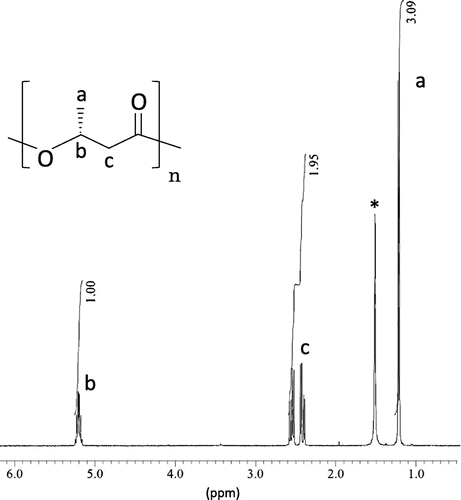
To evaluate PHA accumulation in B. bataviensis JCM 21706T, the strain was cultured in shake flasks containing 100 mL R2A medium (0.5 g peptone, 0.5 g casamino acids, 0.5 g yeast extract, 0.3 g dipotassium phosphate, 0.05 g MgSO4·7H2O, and 20 g glucose per liter of deionized water) on a reciprocal shaker at 150 rpm for 48–72 h at 30 °C and polymer production was evaluated by high-performance liquid chromatography (HPLC) based on crotonic acid determinationCitation6) and gas chromatography (GC) based on methanolysis as previously described.Citation7) The wild-type B. bataviensis strain grown on R2A media produced 0.43 wt% PHA. Once PHA production was verified, one set of potential PHA synthase genes phaRCBb (PhaR and PhaC from B. bataviensis JCM 21706T) among 3 homologs on the genome was identified in the Genbank database (accession number: NZ_AJLS01000000) and amplified by PCR using the following primer set: PhaRCBbF (5′-TCC CTC TAG AAA TAA TTT TGT TTA ACT TTA AGA AGG AGA TAT AAT GGC AGA TCA ACG T-3′) and PhaRCBbR (5′-TTG GTT GGA TCC CTA ATT TTT CCT TGA ACG CGC ACA AAG-3′). The PCR-amplified phaRCBb genes were initially cloned into the pET3a vector (Novagen, Madison, WI, USA) and after sequence verification were subcloned into pGEM′′ABexCitation8) harboring phaAB from Ralstonia eutropha H16 encoding the PHA monomer supplying enzymes. The resulting plasmid (pGEM′′PhaRCBbAB) was then transformed into E. coli JM109 and subsequently stained with Nile red on LB agar plates to visually examine PHA accumulation as previously described.Citation7) To evaluate PHA production from the new class IV PHA synthase genes, recombinant E. coli JM109 was cultured in the lysogeny broth (LB) medium (5 g yeast extract, 10 g peptone, and 10 g NaCl per liter of deionized water) and mixed with of 20 g/L glucose and 100 mg/L ampicillin on a reciprocal shaker at 150 rpm for 72 h at 37 °C. Polymers produced by recombinant E. coli JM109 harboring pGEM”PhaRCBbAB were evaluated in a similar manner as described for B. bataviensis JCM 21706T. The recombinant E. coli expressing phaRCBb accumulated poly(3-hydroxybutyrate) to 5.0 wt%, which was low when compared to the PHA accumulated by recombinant E. coli expressing phaRC genes derived from B. cereus (20–70 wt%).Citation9) The structure of the biosynthesized PHA was confirmed by nuclear magnetic resonance (NMR) analysis with 30 mg purified polymers, solvated in 0.7 mL CDCl3. 1H-NMR spectra shown in Fig. were recorded using a JEOL EC400 spectrometer. The spectrum has the expected chemical shifts of poly(3-hydroxybutyrate) and confirms that the phaRCBb genes encode a functional PHA synthase.
Fig. 2. 1H-NMR spectra of a PHA sample synthesized from glucose by E. coli JM 109 harboring B. bataviensis phaRC genes (pGEM”PhaRCBbAB).
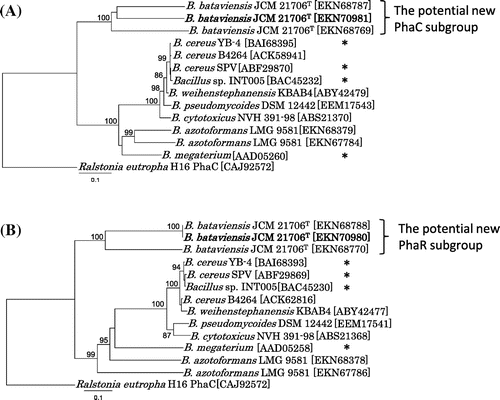
Phylogenetic analyses were performed with the neighbor-joining and Kimura’s method using COBALT software.Citation10) The results revealed a new gene cluster encoding functional PhaRC synthases, which had not been previously identified in any of the previously known PHA synthase groups (Fig. ). The homologies of PhaCBb and PhaRBb to those of B. cereus YB-4Citation11) were only 33 and 19%, respectively. Although no functional motifs for PhaR have been proposed, the deduced molecular weight of PhaRBb was 22.7 kDa, similar to previously described PhaR proteins from B. cereus PhaR (18.5 kDa) and B. megaterium PhaR (22.9 kDa). PhaCBb has a predicted α/β hydrolase domain and a catalytic triad comprised of Cys145, Asp298, and His327 (Fig. S-1). Multiple alignment analysis for PhaCBb demonstrated the typical conserved region known as the PhaC box sequence [(Gly/Ser)-X-Cys-X-(Gly/Ala)-Gly].Citation5) To date, >40 sequences belong to the new PhaC cluster in the Genbank database and it is as large as previously known two subgroups (Fig. S-2). In the new subgroup, phaR was located in the upstream region adjacent to phaC, and no other PHA-related genes were found in the neighboring region; however, in other class IV gene clusters, phaB (acyltransferase) was located between phaR and phaC, and other pha genes (phaP, -Q, and -J) were clustered together (Fig. ).
Fig. 3. Phylogenetic trees of (A) PhaC proteins and (B) PhaR proteins from Bacillus species. B. megaterium,Citation14) B. cereus YB-4,Citation11) INT005,Citation13) and SPVCitation15) marked with asterisks are previously known PhaRC proteins.
Recent studies have reported distinctive features of class IV PHA synthases in which these synthases exhibited alcoholytic cleavage and terminal modification activities to the PHA chain in a manner that is not seen in the other three classes of PHA synthases.Citation12) These features have the potential to expand research in the functionalization of PHA polymers. In addition, potential medical application of these polymers is hampered by the fact that most PHA production takes place in Gram-negative, endotoxin positive strains. Bacillus sp. are Gram-positive and endotoxin negative, which makes them attractive for the production. Further studies on the novel subgroup of class IV synthases in Bacillus will be required for better understanding of catalytic mechanisms of class IV PHA synthases.
Author contributions
Experimental design and interpretation of data were conducted by all authors. K.M., T.K., B.L.R, Z.S, and A.P. carried out experiments. K.M., T.T. and C.T.N. planned the project. K.M. wrote the paper with input from the co-authors, and the other authors reviewed the manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplemental material
The supplemental material for this paper is available at http://dx.doi.org/10.1080/09168451.2016.1230006
TBBB_1230006_Supplementary_Material.pptx
Download MS Power Point (359.5 KB)References
- Sudesh K, Abe H, Doi Y. Synthesis, structure and properties of polyhydroxyalkanoates: biological polyesters. Prog. Polym. Sci. 2000;25:1053–1555.
- Lu J, Tappel RC, Nomura CT. Mini-Review: biosynthesis of poly(hydroxyalkanoates). Polym. Rev. 2009;49:226–248.10.1080/15583720903048243
- Rehm BH. Polyester synthases: natural catalysts for plastics. Biochem. J. 2003;376:15–33.10.1042/bj20031254
- Taguchi S, Ooi T, Mizuno K, et al. Advances and needs for endotoxin-free production strains. Appl. Microbiol. Biotechnol. 2015;99:9349–9360.10.1007/s00253-015-6947-9
- Tsuge T, Hyakutake M, Mizuno K. Class IV polyhydroxyalkanoate (PHA) synthases and PHA-producing Bacillus. Appl. Micrbiol. Biotechnol. 2015;99:6231–6240.10.1007/s00253-015-6777-9
- Karr DB, Waters JK, Emerich DW. Analysis of poly-β-hydroxybutyrate in Rhizobium japonicum bacteroids by ion-exclusion high-pressure liquid chromatography and UV detection. Appl. Environ. Microbiol. 1983;46:1339–1344.
- Ilham M, Nakanomori S, Kihara T, et al. Characterization of polyhydroxyalkanoate synthases from Halomonas sp. O-1 and Halomonas elongata DSM 2581: site-directed mutagenesis and recombinant expression. Polym. Degrad. Stab. 2014;109:416–423.10.1016/j.polymdegradstab.2014.04.024
- Takase K, Taguchi S, Doi Y. Enhanced synthesis of poly (3-hydroxybutyrate) in recombinant Escherichia coli by means of error-prone PCR mutagenesis, saturation mutagenesis, and in vitro recombination of the type II polyhydroxyalkanoate synthase gene. J. Biochem. 2003;133:139–145.10.1093/jb/mvg015
- Tomizawa S, Hyakutake M, Saito Y, et al. Molecular weight change of polyhydroxyalkanoate (PHA) caused by the PhaC subunit of PHA synthase from Bacillus cereus YB-4 in recombinant Escherichia coli. Biomacromolecules. 2011;12:2660–2666.10.1021/bm2004687
- Papadopoulos JS, Agarwala R. COBALT: constraint-based alignment tool for multiple protein sequences. Bioinformatics. 2007;23:1073–1079.10.1093/bioinformatics/btm076
- Mizuno K, Ohta A, Hyakutake M, et al. Isolation of polyhydroxyalkanoate-producing bacteria from a polluted soil and characterization of the isolated strain Bacillus cereus YB-4. Polym. Degrad. Stab. 2010;95:1335–1339.10.1016/j.polymdegradstab.2010.01.033
- Hyakutake M, Tomizawa S, Sugahara I, et al. Carboxy-terminal modification of polyhydroxyalkanoate (PHA) via alcoholysis reaction catalyzed by class IV PHA synthase. Polym. Degrad. Stab. 2015;117:90–96.10.1016/j.polymdegradstab.2015.04.002
- Tajima K, Igari T, Nishimura D, et al. Isolation and characterization of Bacillus sp. INT005 accumulating polyhydroxyalkanoate (PHA) from gas field soil. J. Biosci. Bioeng. 2003;95:77–81.10.1016/S1389-1723(03)80152-4
- McCool GJ, Cannon MC. Polyhydroxyalkanoate inclusion body-associated proteins and coding region in Bacillus megaterium. J. Bacteriol. 1999;181:585–592.
- Valappil SP, Rai R, Bucke C, et al. Polyhydroxyalkanoate biosynthesis in Bacillus cereus SPV under varied limiting conditions and an insight into the biosynthetic genes involved. J. Appl. Microbiol. 2008;104:1624–1635.10.1111/j.1365-2672.2007.03678.x
