Abstract
The Nobel Prize in Physiology or Medicine 2015 was awarded for discoveries related to the control of parasitic diseases using natural products of microbial and plant origin. In current drug discovery programs, synthesized compounds are widely used as a screening source; however, this award reminds us of the importance of natural products. Here, we introduce our phenotypic screening methods based on changes in cell morphology and discuss their effectiveness and impact for natural products in drug discovery.
The first Nobel Prize in Physiology or Medicine was awarded in 1901 to Emil Adolf von Behring (diphtheria antiserum therapy) and the second to Ronald Ross (malaria infection). Thus, combating infectious disease is not only a long-cherished dream of humankind but also the greatest challenge for humankind to keep addressing. To date, there have been numerous Nobel Laureates who worked to elucidate the pathogenesis of diseases and to establish novel therapies; however, only two Prizes have been awarded for discoveries of natural products and their curative effects—Alexander Fleming, Howard Walter Florey, and Ernst Boris Chain (discovery of penicillin) in 1945 and Selman Abraham Waksman (discovery of streptomycin) in 1952. Most recently, the Nobel Prize in Physiology or Medicine 2015 was awarded to Satoshi Ōmura and William Cecil Campbell for their discoveries concerning a novel therapy against infections caused by roundworm parasites (ivermectin) and to Youyou Tu for discoveries concerning a drug of plant origin (artemisinin) as a novel therapy against malaria.
The period from the 1950s through the 1970s was the Gold Rush era for exploration of novel nature-derived antibiotics; new substances derived from cultured broth of microbes or plant extracts led to a string of drugs. Soon afterward, the researchers who discovered the potential in natural products utilized them as screening sources for anticancer agents, immunosuppressors, inhibitors of cholesterol biosynthesis, and antibiotics. Since fermentation (miso, natto) and brewing (Japanese sake) are traditionally a Japanese forte, Japan has produced numerous giants in agricultural chemistry: Yusuke Sumiki (gibberellin, blastcidin S), Hamao Umezawa (kanamycin, bleomycin), Tojyu Hata (leucomycin, mitomycin C), and Akira Endo (mevastatin).
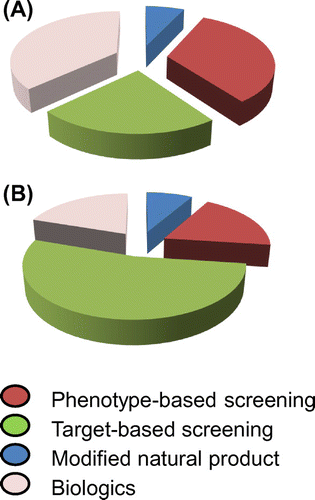
After the 1980s, drug discovery from natural products became difficult, and high-throughput screening (HTS) using chemically synthesized compounds flourished. Large pharmaceutical companies have constructed chemical libraries with millions of compounds and conducted large-scale screening campaigns. However, we wonder whether synthetic chemicals are favorable sources for the discovery of new drugs and whether careful phenotypic evaluation of a limited number of samples (high-content screening; HCS) is already outdated. Interestingly, Swinney and Anthony reported that during the 10-year period between 1999 and 2008, many first-in-class drugs were discovered through phenotypic screening, while many me-too drugs (new drugs without novelty) were discovered through target-based screening (Fig. ).Citation1)
Bioassay system based on changes in cell morphology
To screen for bioactive small molecules, it is essential to establish a simple bioassay system. Screening systems are roughly categorized into enzyme-, cell-, and animal-based assays, each with its own pros and cons. When constructing a new bioassay system, we make a point of verifying whether it has the 4S (Simple, Speedy, Sensitive, and Specific) and 1D (Distinctive) framework. In other words, a good screening system comprehends an easy-to-follow assay protocol, a quick turn-around time for assay results, high sensitivity, high specificity, and easy interpretation of assay results. Even though cell-based assays do not always meet the 4S & D criteria, we feel that such phenotypic screening and natural products are compatible.
Screening for antifungal compounds
There are many infectious fungi such as human-pathogenic Aspergillus fumigatus and phytopathogenic Pyricularia oryzae. Until now, several antifungal drugs such as amphotericin, micafungin, and polyoxin have been developed for medical or agrochemical use; however, they have undesirable adverse effects and led to the emergence of drug-resistant strains. Therefore, a tireless effort to develop new antifungal drugs is needed. Because fungi are eukaryotes and have high biochemical and cytological similarities with their hosts, it is difficult to identify effective molecular targets. Indeed, almost all potent antifungal agents are the modulators of fungi-specific cell wall biosynthesis.
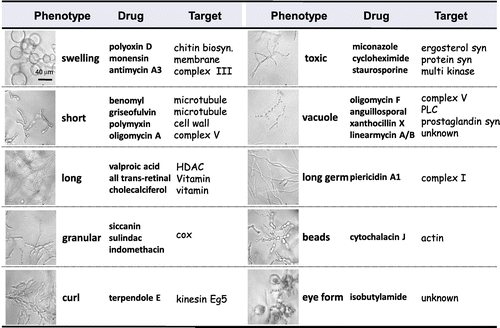
Previously, we isolated polyoxin from one actinomycete and successfully applied it for practical use as an agricultural fungicide.Citation2) Endo and colleagues demonstrated the detailed mode of action of polyoxin—inhibition of cell wall synthesis of filamentous fungi—and that this drug accordingly causes a unique cell-shape change, “swelling.”Citation3) Based on previous work,Citation4) we constructed a database of morphological changes in the rice blast fungus P. oryzae and utilized it for the screening of novel antifungal agents.
Among filamentous fungi, P. oryzae is known to possess a particularly large number of extracellular signal receptors and highly respond to chemicals, which induce various morphological changes. We have comprehensively examined the effects of various well-characterized agents on cell-shape change in P. oryzae in a time- and dose-dependent manner and investigated the “drug-target-phenotype” relationships. Our results indicated that the characteristic morphological changes are classified, based on the morphological features and the mechanism action, into 10 phenotypes: “swelling,” “short,” “granular,” “toxic,” “long,” “vacuolation,” “long germ,” “beads,” “eye form,” and “curl” (Fig. ). This encyclopedic morphology database allows us to easily discriminate among the phenotypes induced by well-known antifungals, such as polyoxins (chitin synthesis; swelling), azoles (ergosterol synthesis; toxic), and griseofulvin (tubulin; short).
Next, we performed HCS using over 5000 compounds deposited in a chemical library of RIKEN NPDepo and ca. 7200 cultured broths as test samples. We identified a cyclic depsipeptide, lipopeptin,Citation5) and a chlorine-containing shikimic acid derivative,Citation4) both of which were reported to induce unique morphological changes, suggesting that our database is highly effective.
Screening for anticancer agents
When screening for antifungal agents, we visually observe the fungal morphology; however, for screening using mammalian cells, an automatic microscope system (IN Cell Analyzer, CellVoyager, CellInsight, etc.) is now available. With the multi-parameter image processing and visualization tools, both the qualification and quantification of various cellular phenotypes, such as morphological changes, can be realized in a “high-throughput” fashion.
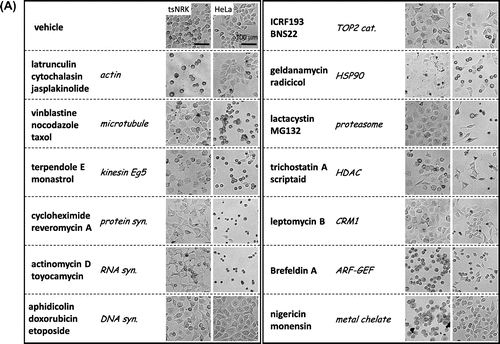
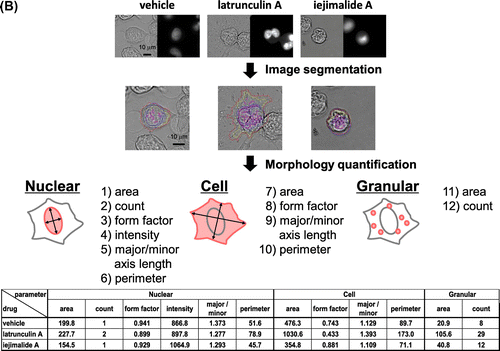
As for fungi, we found that mammalian cancer cells undergo specific morphological changes related to the mode of action of a drug (Fig. (A)). Thus, we aimed to construct a phenotypic screening system based on the high-content cell morphological database, “MorphoBase.” After several experiments using various cell lines, we decided to use src-tsNRK and HeLa cells for the assay system.Citation6,7) Both cell lines exhibited a variety of changes in shape in response to various compounds. To eliminate unintentional human errors during visual observation, we performed automated observations using IN Cell Analyzer (Fig. (B)). Specifically, we designed a high-content imaging algorithm that segments an individual cell into three fragments—“Cell,” “Nuclear,” and “Granular”—and quantifies 12 morphological parameters. Furthermore, we developed a data analysis program that incorporates multivariate statistical tools. Finally, this system can classify the mode of action of typical anticancer drugs and profile various pharmacological targets of mechanistically unknown compounds and even crude extracts.Citation8)
Fig. 3. Screening system indexed by morphological changes in cancer cells.
Pyrrolizilactone is a novel fungal metabolite, which has a unique chemical structure and induces a specific morphological change.Citation9) Phenotypic profiling based on MorphoBase (morphological changes) and ChemoProteoBase (proteome variance) demonstrated that pyrrolizilactone is associated with proteasome inhibition. Subsequent biochemical experiments established that pyrrolizilactone inhibits the 20S proteasome activities, especially the trypsin-like activity.Citation10)
Summary
In the screening for bioactive compounds, HCS based on careful observation of cell morphology is lower throughput yet often more efficient and powerful than HTS. Consistently, our morphology-based strategy readily classifies the phenotypes of authentic compounds by their modes of action and predicts the targets of a compound of interest; this method will facilitate the discovery of novel drug candidates from natural products.
The Nobel Prize in Physiology or Medicine 2015 will increase the allure of natural products and calls for our further efforts with ingenuity to achieve further breakthrough innovations in natural products.
Disclosure statement
No potential conflict of interest was reported by the authors.
Funding
This work was supported in part by a Grant-in-Aid for Scientific Research (A) from the Ministry of Education, Culture, Sports and Technology of Japan [grant number 15H02445], the Science and Technology Research Promotion Program for Agriculture, Forestry, Fisheries, and Food Industry, and the Research Program on Hepatitis from the Japan Agency for Medical Research and Development, AMED.
Notes
† This work is a translation of an original work in Japanese in the Japan Society for Bioscience, Biotechnology, and Agrochemistry http://doi.10.1271/kagakutoseibutsu.54.32
References
- Swinney DC, Anthony J. Nat Rev Drug Discov. 2011;10:507–519.10.1038/nrd3480
- Suzuki S, Isono K, Nagatsu J, et al. J Antibiot (Tokyo). 1965;18:131.
- Endo A, Kakiki K, Misato T. J. Bacteriol. 1970;104:189–196.
- Kitamura E, Hirota A, Nakagawa M, et al. Tetrahedron Lett. 1990;31:4605–4608.10.1016/S0040-4039(00)97687-1
- Tsuda K, Kihara T, Nishii M, et al. J Antibiot (Tokyo). 1980;33:247–248.10.7164/antibiotics.33.247
- Osada H, Koshino H, Isono K, et al. J Antibiot (Tokyo). 1991;44:259–261.10.7164/antibiotics.44.259
- Muroi M, Kazami S, Noda K, et al. Chem Biol. 2010;17:460–470.10.1016/j.chembiol.2010.03.016
- Futamura Y, Kawatani M, Kazami S, et al. Chem. Biol. 2012;19:1630–1640.
- Nogawa T, Kawatani M, Uramoto M, et al. J Antibiot (Tokyo). 2013;66:621–623.10.1038/ja.2013.55
- Futamura Y, Kawatani M, Muroi M, et al. ChemBioChem. 2013;14:2456–2463.10.1002/cbic.v14.18