Abstract
Topmouth culter (C. alburnus) is an important commercial fish in China. We compared the nucleotide variations in the mtDNA genomes among three geographical groups of Culter alburnus: Liangzi Lake, Hubei Province (referred to as LZH); Taihu Lake, Jiangsu Province (TH); and Poyang Lake, Jiangxi Province (PYH). The similarity of whole mtDNA genomes ranged from 0.992 to 0.999. The similarity among 13 protein-coding genes, 2 rRNA genes, and the D-loop sequences was found to range from 0.982 to 0.996. This is useful data for future designing work for making specific molecular marker for distinguishing individuals of C. alburnus from the three geographical groups. An extended termination-associated sequence (ETAS) and several conserved blocks (CSB-F, CSB-E, CSB-D, CSB1, CSB2, and CSB3) were identified in the mtDNA control regions. A phylogenetic analysis shows a monophyletic relationship of the LZF-female and the LZF-male. However, the analysis also showed paraphyletic relationships for the other two geological groups. This result will be useful for the future breeding work of C. alburnus.
Graphical abstract
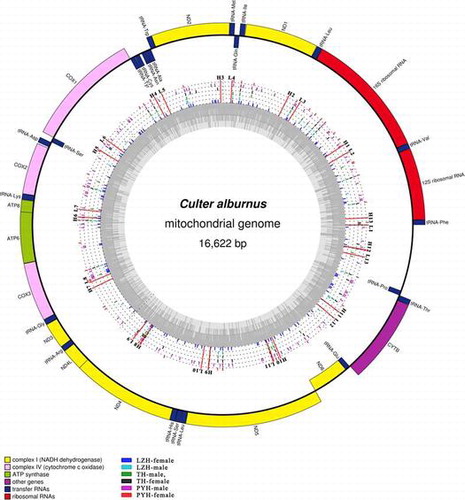
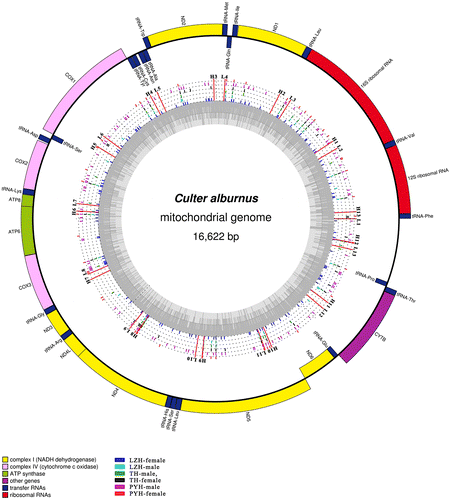
Circular representation of the complete mitochondrial genomes of C. alburnus.
Topmouth culter (C. alburnus) is widely distributed in major reservoirs, rivers, and lakes of ChinaCitation1). With increasing annual cultured production, C. alburnus has already become an important commercial fish in ChinaCitation2,3). However, some production units have not paid sufficient attention to resource management during the aquaculture production process, causing germplasm degradation phenomena including slow growth and increased disease among farmed fishCitation4). The production of C. alburnus does not currently meet the consumer’s demand because of the sharp decline in the C. alburnus’ resourcesCitation5). Germplasm resources have become the main constraint factor for the stable development of C. alburnus. Therefore, it is urgent to cultivate at least one high-yield, fast-growing, and highly disease-resistant varietyCitation6).
At present, we conducted a genetic breeding program with other colleagues for selective breeding and to prevent the genetic degeneration of C. alburnus. Based on the initial groups from three geographic regions, the family construction technology of C. alburnus was studied with directional mating using 1 female and 1 male. From the 12 mated pairs, 8 full-sib families were constructed, and 15,000 individuals of the F1 generation were obtained. The C. alburnus populations from the three geographic regions are morphologically and physiologically similar. The traditional morphological identification method is not suitable for distinguishing C. alburnus. MtDNA is maternally inherited, and subsequent generations would only have the maternal DNACitation7). Therefore, mtDNA sequences can be used as a supplementary means to distinguish the three geographical C. alburnus groupsCitation8,9). In this study, we reconstructed the complete mtDNA sequences of C. alburnus from three geographical groups. With the comparative analyses of those six mtDNA, part of the genes could be selected as specific molecular markers for distinguishing three geographical C. alburnus groups. These specific molecular markers can also be used to evaluate the genetic stability between the F1 generation and their parents.
Materials and methods
Sample collection
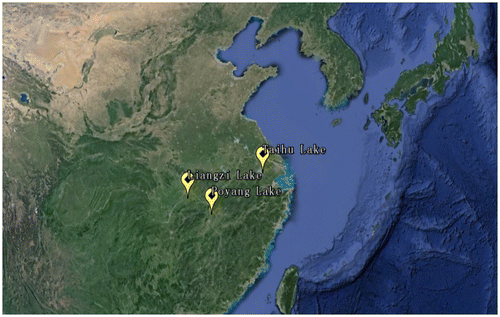
C. alburnus individuals were collected from following three geographic regions (Fig. ); Liangzi Lake on Hubei Province (LZH), Taihu Lake on Jiangsu Province (TH), and Poyang Lake on Jiangxi Province(PYH). Poyang Lake (28°24′–29°46′N, 115°49′–116°46′E), China’s largest freshwater lake, is located on the south bank of the middle and lower reaches of Yangtze River. Taihu Lake (30°55′–31°32′N, 119°52′–120°36′E), the third largest freshwater lake in China, is located in the Yangtze River Delta and is approximately 100 km2 away from the metropolis of Shanghai. Liangzi Lake (30°03′–30°20′N, 114°26′–114°38′E) is a typical shallow macrophytic lake in the middle reach of Yangtze River in Hubei Province, China. The C. alburnus resources were abundant in Poyang Lake, Liangzi Lake, and Taihu Lake.
DNA extraction
Genomic DNA was isolated from the tail fin of fish by using the Universal Genomic DNA Extraction Kit (Takara, Japan). All C. alburnus individuals were released after sampling. All reasonable efforts were made to minimize suffering and the number of animals used. DNA quality was assessed using 1% agarose gel electrophoresis. DNA concentration was measured on a NanoDrop 2000 Spectrophotometer (Thermo Scientific) at 260 nm.
Polymerase chain reaction and sequencing
The 13 pairs of primers (Table ) for amplifying the mtDNA sequences of the three geographical groups were designed based on the complete mitochondrial genome of Xingkai topmouth culter (KF039719) in GenBankCitation10). The PCR program was as follows: denaturation at 94 °C for 3 min; followed by 35 cycles of 94 °C for 30 s, 56 °C for 30 s, and 72 °C for 30 s; and a final extension at 72 °C for 10 min. The reaction was carried out in a volume of 25 μL containing 100 ng genomic DNA, 2.5 μL of 10 × buffer, 0.4 μmol/L primer (each direction), 2 μmol/L dNTP (each), and 1 unit of TaqDNA polymerase (Takara, Dalian, China). The amplified fragments were subcloned into the vector pMD19-T (Takara, Japan) and sequenced.
Table 1. PCR primers in the analysis of the C. alburnus mitochondrial genomes.
Data and sequence analysis
All of the primers were designed using Premier 5.0 and Oligo 6.0 software. Sequences were edited and aligned using ClustalX 1.83, and manually adjusted to minimize mismatchesCitation11). Phylogenetic analysis was performed using the MEGA4.1 software by bootstrap analysis.
Results and discussion
Genome organization
The total lengths of the C. alburnus mitogenomes were determined to be 16,620 bp for LZH female (GenBank accession number KX829023), 16,620 bp for LZH male (KX829024), 16,622 bp for TH female (KX829025), 16,622 bp for TH male (KX829026), 16,622 bp for PYH female (KX829027), and 16,622 bp for PYH male (KX829028). The complete mitochondrial genome sequence of Xingkai topmouth culter (Culter alburnus) was determined to be 16,622 bp in lengthCitation10). Further, the size of the mitochondrial genome from these three populations is approximately the same as that of previously published mtDNA of this fish Citation10).
The gene arrangement was identical among the three geographical groups (Fig. ). We confirmed that our data are consistent with previously published data for the mitochondrial genome of Xingkai topmouth culter (Culter alburnus)Citation10). Six mtDNA genomes encode 13 protein subunits, 22 tRNAs, and two rRNAs. All genes were encoded on the H-strand, except for the Nad6 and the eight tRNA genes (Gln, Ala, Asn, Cys, Tyr, Glu, Pro, and Ser), which were encoded on the L-strand. Nearly all of the protein-coding genes used ATG as the start codon. The only exception was the Cox1 gene, which used GTG instead of ATG as a start codon. Nine protein-coding genes in C. alburnus ended with complete stop codons TAA (Nad1, Cox1, Atp6, Cox3, Nad4l, and Nad5) or TAG (Nad2, Atp8, and Nad6). We found that two protein-coding genes (Nad3 and Nad4) had the incomplete stop codon TA, and the other 2 genes ended with the incomplete stop codon T (Cox2 and Cob). These results were the same as for Culter compressocorpusCitation12) and Culter recurvicepsCitation13).
Fig. 2. Circular representation of the complete mitochondrial genomes of C. alburnus. Circles display (from inside to outside): LZH-female, LZH-male, TH-male, TH-female, PYH-male, and PYH-female. Different color rods and dots represent the variable sites and the same nucleotides, respectively. L1–L13: the location of forward primers. H1–H13: the location of reverse Primer.
Similarity and divergence analysis
As shown in the Table , the overall genome sequence similarity (ratio) ranged from 0.992 to 0.999 among the three geographical groups (Supplemental 1). The highest sequence similarity was observed between LZH female and LZH male (0.999). The low sequence similarity was observed between LZH female and TH female (0.993), LZH female and TH male (0.993), LZH female and PYH female (0.993), LZH female and PYH male (0.992), LZH male and TH female (0.993), LZH male and TH male (0.994), LZH male and PYH female (0.993), and LZH male and PYH male (0.993). These results could be used as a reference for selecting appropriate parents for the breeding work.
Table 2. Similarity of 16 homolog genes among C. alburnus (retention three significant digits).
The sequence similarity of 13 homologous genes ranged from 0.982 to 1.00 (Table ). We listed the loci among the 3 geographical groups for the 13 orthologous genes where allele frequency is high as follows: LZH female and LZH male (Nad2, 0.998), LZH female and TH female (Nad6, 0.984; Cob, 0.984), LZH female and TH male (Nad6, 0.984; Cob, 0.984), LZH female and PYH female (Nad6, 0.984; cob, 0.984), LZH female and PYH male (Cob, 0.982), LZH male and TH female (Nad6, 0.984; Cob, 0.984), LZH male and TH male (Nad6, 0.984; Cob, 0.984), LZH male and PYH female (Nad6, 0.984; Cob, 0.984), LZH male and PYH male (Cob, 0.982), TH female and TH male (Nad4, 0.994), TH female and PYH female (D-loop, 0.995), TH female and PYH male (D-loop, 0.996), TH male and PYH female (Nad4, 0.994), TH-male and PYH-male (Nad3, 0.994), PYH-female and PYH-male (D-loop, 0.994). Based on the listed gene sequences, specific molecular markers can be designed for distinguishing the three geographical groups of C. alburnus.
The differences in particular genes are related to evolution. This study identified inconsistent evolutionary relationships based on the similarity of the whole genomes and of particular genes. For example, although the similarity in the genome between LZH male and TH female (0.993) is lower than that between TH male and PYH male (0.998), the similarity in the gene Nad1 for the former pair (0.998) is greater than that for the latter pair (0.995). The literature has demonstrated that the complete mtDNA sequences of eight teleosts could be used to reproduce the expected phylogeny with high statistical support, unlike the sequences of individual genesCitation14). In this study, individual mitochondrial genes might potentially suggest misleading evolutionary relationships and might not be representative of the entire mitochondrial genome. Inconsistent results have also been reported in some previous studiesCitation15,16). In this study, the species evolutionary relationship among the three geographic groups varied considerably among the various mitochondrial genes. The different rates of evolution of mitochondrial genes among the three geographic groups may contribute to these inconsistent results.
Ribosomal RNA genes and Transfer RNA genes
In all three geographical groups, the 12S rRNA gene has the same length, 952 bp, and is located between the tRNA-Phe and tRNA-Val genes. The sequence similarity of 12S rRNA genes ranged from 0.989 to 0.997. The 16S rRNA gene, with lengths of 1693 bp for LZH female, 1693 bp for LZH male, 1692 bp for TH female, 1692 bp for TH male, 1693 bp for PYH female, and 1692 bp for PYH male, is located between the tRNA-Val and tRNA-Leu genes. The sequence similarity of the 16S rRNA genes was found to be between 0.995 and 0.999.
In total, 22 tRNA genes of C. alburnus have been determined. They are interspersed between the rRNA and protein-coding genes, ranging in size from 67 to 76 bp, similar to the cases of Culter dabryiCitation17) and Culter recurvicepsCitation13). The differences in the 22 tRNA genes among the three geographical groups were as follows: one base in tRNA-Trp from LZH female differed from the other 5 groups, one base in tRNA-Asn of PYH female differed from the other 5 groups, one same base in tRNA-Gly of LZH female and LZH male differed from the other 4 groups, and one same base in tRNA-Pro of LZH female and LZH male differed from the other 4 groups.
Non-coding regions
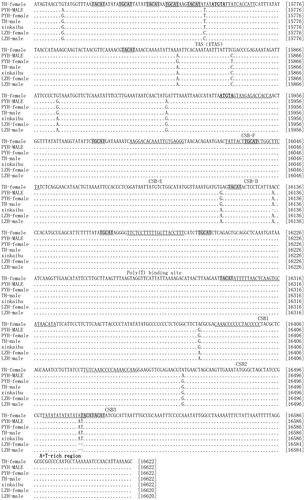
As in other vertebrates, the C.alburnus also has a control region located between tRNA-Pro and tRNA-Phe within their mitochondrial genomesCitation10) (Fig. ). One extended termination-associated sequence (ETAS) is recognized in these fishes. The consensus sequence of the ETAS in some fishes is TACATAT–ATGTATT–ATCACCATATATTATATTAACCAT with the motif TACATCitation18). The sequence of ETAS in Culter fishes is TGCATAAGTACATATATATGTATTATCACCATT, with the motif sequences TGCAT and TACAT, and one palindromic sequence, ATGTA. This sequence is known as the motif which is associated with the termination of D-loop strand synthesisCitation19). Three central conserved sequence blocks (CSB-F, CSB-E, and CSB-D) have been detected in Culter fishes. The consensus sequence of CSB-F, CSB-E, and CSB-D in Culter fishes was highly conserved and was consistent with those described for other fish speciesCitation18). CSB1 was associated with the initiation of mtDNA duplication. The consensus sequence of CSB1 in the Culter fishes was recognized as ATTTTTAACTCAAGTGCATAACATA, and the highly conserved sequence was TGCATA. Another two conserved sequence blocks, CSB2 (CAAACCCCCCTACCCCC) and CSB3 (TGTCAAACCCCAAAACCAAG), were highly conserved within the Culter fishes and were consistent with the CSB2 and CSB3 identified in some other fishesCitation18). The CSB sequences are associated with mitochondrial DNA replication and transcriptionCitation20).
Fig. 3. The complete control regions of three geographical groups. Termination associated sequences (TAS) as well as conserved sequence blocks (CSB-D, CSB-E, CSB-F CSB-1, CSB-2, and CSB-3) were underlined. Dots and dashes indicated the same nucleotides and gaps, respectively. Shaded region designated the motif of TGCAT and TACAT. ATGTA were bold. The number on the right side of alignment shows the nucleotide positions.
An additional non-coding region, the L-strand replication origin (OL), was located in a cluster of tRNA-Trp, tRNA-Ala, tRNA-Asn, tRNA-Cys, and tRNA-Tyr (the WANCY region). The sequences of the three geographical groups were identical in this 46 bp segment, 5′-GCCGGGGCTTTTCCCGCCTTACTAGGCTTTAGGCGGGAAAAGTAGA-3′. These sequences have the potential to form a stable stem-loop structureCitation21).
Phylogenetic analyses
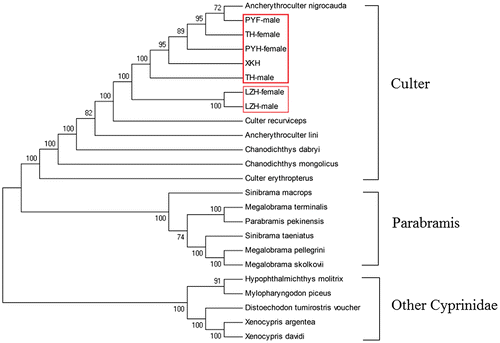
As shown in Fig. , there were three independent clusters in the phylogenetic tree. All Culter fish, including Culter alburnus (XKH), Ancherythroculter nigrocauda, Culter recurviceps, Chanodichthys dabryi, Chanodichthys mongolicus, Ancherythroculter lini, and Culter erythropterus, were clustered at one branch in the phylogenetic tree. The other cyprinid fish species were distributed in two other branches. These results showed that the Culter fishes are monophyletic. In addition, LZH female and LZH male are located together in a branch; however, other five groups, TH-male, XKH, PYH-female, PYH-male, TH-female create another monophyletic clade with Ancherythroculter nigrocauda. These results suggested that the evolutionary relationship between the LZH female and the LZH male is close, particularly in comparison to the relationship between the LZH female and LZH male and the other culter fishes, which is more distant. This result has important significance for the breeding of C. alburnus. It suggests that crossbreeding work may be impossible for LZH females and LZH males’ combinations of TH males, PYH females, PYH males, and TH females. Improved varieties of C. alburnus should be cultivated by breeding among different geographical groups.
Fig. 4. Phylogenetic analysis based on the complete mitochondrial DNA sequences of various fish. Numbers at tree nodes refers to percent bootstrap values after 1000 replicates. The LZH-female, LZH-male, TH-female, TH-male, PYH-female, and PYH-male were boxed in the phylogenetic tree. Culter alburnus (XKH, KF039719.1), Ancherythroculter lini (KR864861.1), Ancherythroculter nigrocauda (KC513573.1), Chanodichthys dabryi (KC526217.1), Chanodichthys mongolicus (KC701385.1), Culter erythropterus (KJ801524.1), Culter recurviceps (KJ609181.1), Distoechodon tumirostris voucher (DQ026431.1), Hypophthalmichthys molitrix (KJ746944.1), Megalobrama pellegrini (KP262030.1), Megalobrama skolkovii (KJ630486.1), Megalobrama terminalis (AB626850.1), Mylopharyngodon piceus (EU979305.1), Parabramis pekinensis (JX242531.1), Sinibrama macrops (KC122916.1), Sinibrama taeniatus (KM272589.1), Xenocypris argentea (AP009059.1), Xenocypris davidi (KF039718.1).
Author contributions
Jianwu Shi and Yijiang Hong conceived the experiments. Jianwu Shi, Dexia Wang, Junqing sheng, Peng Kou, and Beijuan Hu designed and performed the experiments. Jianwu Shi analyzed the data. Junhua Wang contributed reagents and analysis tools. Liugen Zeng and Minghe Xiao contributed the experiment materials. Jianwu Shi and Yijiang Hong wrote the paper.
Funding
This work was supported by State Science and Technology Support Program of China [grant number 2012BAD25B06]; the earmarked fund for Jiangxi Agriculture Research System [grant number JXARS-10], [JXARS-02]; the open fund of Key Laboratory of Poyang Lake Environment and Resource Utilization in Nanchang University [PYH2015-14]; the Key Scientific and Technological Program of Jiangxi Province, China [grant numbers. 20152ACF60013], [KJLD12001], [2009BNA07400].
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplemental material
Supplemental material for this article can be accessed at http://dx.doi.org/10.1080/09168451.2016.1270739.
TBBB_1270739_Supplementary_Material.xlsx
Download MS Excel (23.5 KB)References
- Luo Y, Chen Y. Cultrinae. In: Chen YY, Chu XL, Luo YL, et al., editors. Fauna Sinica. Osteichthyes. Cypriniformes II. Beijing: Science Press; 1998. p. 112–207.
- Wang W, Chen L, Yang P, et al. Assessing genetic diversity of populations of topmouth culter (Culter alburnus) in China using AFLP markers. Biochem Syst Ecol. 2007;35:662–669.10.1016/j.bse.2007.04.008
- Qi PZ, Guo BY, Xie CX, et al. Assessing the genetic diversity and population structure of Culter alburnus in China based on mitochondrial 16S rRNA and COI gene sequences. Biochem Syst Ecol. 2013;50:390–396.10.1016/j.bse.2013.04.010
- Qi PZ, Qin JH, Xie CX. Determination of genetic diversity of wild and cultured topmouth culter (Culter alburnus) inhabiting China using mitochondrial DNA and microsatellites. Biochem Syst Ecol. 2015;61:232–239.10.1016/j.bse.2015.06.023
- Hu TJ, Zhou ZM, Zhang WM, et al. Biology and breeding technology of topmouth culter, Culter alburnus. Water Fish. 2003;23:20–21.
- Lei S, Chen X, Wang J, et al. An integrated and comprehensive transcriptome reveals immune-related genes and signal pathways in topmouth culter (Culter alburnus). Aquac Res. 2016. doi: 10.1111/are.13060.
- John JCS, Facucho-Oliveira J, Jiang Y, et al. Mitochondrial DNA transmission, replication and inheritance: a journey from the gamete through the embryo and into offspring and embryonic stem cells. Hum Reprod Update. 2010;16:488–509.10.1093/humupd/dmq002
- Witt JDS, Threloff DL, Hebert PDN. DNA barcoding reveals extraordinary cryptic diversity in an amphipod genus: implications for desert spring conservation. Mol Ecol. 2006;15:3073–3082.10.1111/mec.2006.15.issue-10
- Ward RD, Zemlak TS, Innes BH, et al. DNA barcoding Australia’s fish species. Philos T R Soc B. 2005;360:1847–1857.10.1098/rstb.2005.1716
- Liu Y, Yang J. The complete mitochondrial genome sequence of Xingkai topmouth culter (Culter alburnus). Mitochondr DNA. 2014;25:451–453.10.3109/19401736.2013.809430
- Thompson JD, Gibson TJ, Plewniak F, et al. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882.10.1093/nar/25.24.4876
- Xu M, Tong GX, Geng LW, et al. Mitochondrial DNA sequence of Culter compressocorpus. Mitochondr DNA. 2016;27:643–644.10.3109/19401736.2014.908470
- Yang H, Zhao H, Sun J, et al. The complete mitochondrial genome of the Culter recurviceps (Teleostei, Cyprinidae). Mitochondr DNA. 2016;27:762–763.10.3109/19401736.2014.915529
- Miya M, Nishida M. Use of mitogenomic information in teleostean molecular phylogenetics: a tree-based exploration under the maximum-parsimony optimality criterion. Mol Biol Evol. 2000;17:437–455.
- Cao Y, Adachi J, Janke A, et al. Phylogenetic relationships among eutherian orders estimated from inferred sequences of mitochondrial proteins: instability of a tree based on a single gene. J Mol Evol. 1994;39:519–527.
- Zardoya R, Meyer A. Phylogenetic performance of mitochondrial protein-coding genes in resolving relationships among vertebrates. Mol Biol Evol. 1996;13:933–942.10.1093/oxfordjournals.molbev.a025661
- Zhang X, Ma Z, Chen D, et al. The complete mitochondrial genome of Culter dabryi (Cyprinidae: Cultrinae). Mitochondr DNA. 2014;25:98–99.10.3109/19401736.2013.784754
- Liu HZ. The structure and evolution of the mtDNA control region in fish: taking example for Acheilognathinae. Prog Nat Sci. 2002;12:266–270 . (in Chinese with an English abstract).
- Broughton RE, Milam JE, Roe BA. The complete sequence of the zebrafish (Danio rerio) mitochondrial genome and evolutionary patterns in vertebrate mitochondrial DNA. Genome Res. 2001;11:1958–1967.
- Zhao JL, Wang WW, Li SF, et al. Structure of the mitochondrial DNA control region of the sinipercine fishes and their phylogenetic relationship. Acta. Genet Sin. 2006;33:793–799.10.1016/S0379-4172(06)60112-1
- Cheng Y, Wang R, Sun Y, et al. The complete mitochondrial genome of the small yellow croaker and partitioned Bayesian analysis of Sciaenidae fish phylogeny. Genet Mol Biol. 2012;35:191–199.10.1590/S1415-47572012005000006