Abstract
GDP-d-mannose pyrophosphorylase (GMP) is one of the enzymes that highly expressed in acerola plants. A promoter assay suggests the presence of a new cis-element in the −1087 to −1083 bp sequence of the MgGMP promoter. Moreover, cis-elements, present in the −1080 to −600 bp sequence of the MgGMP promoter, function as enhancers of MgGMP expression.
Ascorbic acid (AsA) is an essential material for plant growth. It plays a crucial role in reactive oxygen species scavenging because it is the most abundant cellular antioxidant,Citation1) and it also plays a role in photosynthesis, phytohormone biosynthesis, and flowering.Citation2–4) GDP-d-mannose pyrophosphorylase (GMP) catalyzes the conversion of d-mannose 1-phosphate to GDP-d-mannose, the first step in the major AsA biosynthetic pathway of plants.Citation5) In Arabidopsis, the GMP-defective mutant vtc1 contains 25% of the wild-type levels of AsA.Citation6) Moreover, levels of AsA can be increased to more than twice of those in wild-type tobacco plants by overexpressing tomato GMP.Citation7) Arabidopsis GMP (AtGMP) is regulated at the transcriptional level by ethylene response factor 98 acting as an activator.Citation8)
Acerola (Malpighia glabra) contains high levels of AsA in its fruits. In our previous study, we suggested that the high transcriptional expression levels of enzymes involved in the major AsA biosynthetic pathway contributed to the high accumulation of AsA in acerola.Citation9) Acerola GMP (MgGMP) is one of the enzymes that highly expressed in acerola fruits and leaves. Based on promoter assays using protoplasts of tobacco BY2 cells, the essential cis-acting element could be located between −484 and −317 bp upstream from the transcription start site of MgGMP.Citation10) However, the MgGMP promoter had only a 2.6-fold higher activity than the AtGMP promoter, whereas quantitative RT-PCR showed that acerola leaves had 100-fold higher GMP expression levels than Arabidopsis leaves. Therefore, we could not exclude the possibility that other unknown elements in the MgGMP promoter were responsible for the high gene expression.
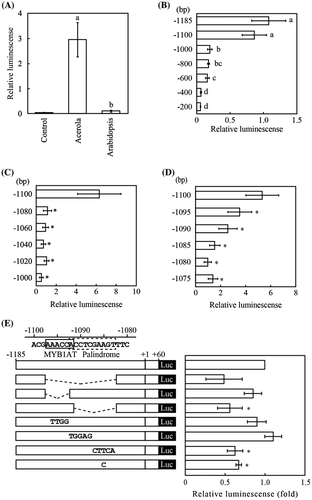
In the present study, we first re-evaluated MgGMP promoter activity using Arabidopsis mesophyll protoplasts by applying the method used by Suekawa et al.Citation11), with a slight modification. The transient transfection of Arabidopsis protoplasts and the assay of luciferase activity were performed as follows.Citation12) The pMONT vector was used as an internal control vector against the Renilla luciferase experimental vector.Citation13) The DNA fragment of the 5′-upstream region (−1185 – + 60 bp) of the MgGMP gene was fused to a reporter gene encoding Renilla luciferase and introduced into Arabidopsis protoplasts. As shown in Fig. (A), 1185 bp of the 5′-upstream region from the transcription start site of MgGMP showed luciferase activities that were approximately 28-fold higher than those of the 5′-upstream region (1200 bp from the initiation codon) of AtGMP. Because 1185 bp of the 5′-upstream region of the MgGMP gene showed high promoter activity using Arabidopsis protoplasts, we further examined the promoter activity using a series of 5′-end deletion constructs of the MgGMP promoter. Deletion analysis showed that decreasing the size of the promoter region to 1100 bp did not considerably alter luciferase activity (Fig. (B)). However, a marked decrease in luciferase activity was observed when the size was shortened to 1000 bp. In addition, there was a significant decrease in luciferase activity when the 5′-upstream region of the MgGMP promoter was deleted to −400 from −600 bp. Although, the latter finding is in agreement with the previous results suggesting that an enhancer is present between −484 and −317 bp of the MgGMP promoter,Citation10) other unknown enhancers appear to be present between −1100 and −1000 bp of the MgGMP promoter. Therefore, we examined promoter activities using the second set of deletion constructs (deleted successively by 20 bp) between −1100 and −1000 bp of the MgGMP promoter. The deletion of the 5′-upstream region down to −1080 bp resulted in a drastic decrease in the luciferase activity (Fig. (C)). Promoter analysis using further fine deletion constructs showed that luciferase activities gradually decreased in line with a progressive increase in 5′-deletion down to 1075 bp (Fig. (D)). The results suggest that potent enhancer(s) are present between −1100 and −1080 bp of the MgGMP promoter. From sequence analysis of the MgGMP promoter using the PLACE program,Citation14) a sequence similar to the MYB1AT element (WAACCACitation15)) was found to be located between −1097 and −1092 bp of the MgGMP promoter. In addition, a palindrome-like sequence (ACCTCGAAGT at −1092 to −1083 bp) was found in the region. Thus, to aid further investigation within the region of the MgGMP promoter, three internal deletion constructs were first tested: from −1097 to −1083 bp, from −1097 to −1092 bp, and from −1091 to −1083 bp (Fig. (E)). As the result of the deletion analysis, the internal deletion from −1083 to −1097 bp showed a tendency to reduce luciferase activity. And the internal deletion from −1083 to −1091 bp caused a significant reduction in luciferase activity, although the internal deletion from −1092 to −1097 bp resulted in no significant reduction in luciferase activity. Following this, the sequence from −1097 to −1083 bp was replaced with its complementary sequences at 4- and 5-bp intervals. No significant difference in luciferase activity was observed when the sequences of −1096 to −1093 bp and −1092 to −1088 bp were replaced with TTGG and TGGAG, respectively. However, a significant reduction was observed with replacement of the sequence from −1087 to −1083 bp. Therefore, the results suggest that the MYB1AT and the palindrome-like sequences do not serve as regulators of MgGMP gene expression, whereas the sequence (GAAGT) from −1087 to −1083 bp functions as an enhancer of MgGMP expression. After careful review, we found GAAGT to be an abscisic acid response element (ABRE)-like sequence. The ABRE consensus sequence is commonly known as CACGTG (ACGT; core sequence). Sarkar and LahiriCitation16) reported that the sequence of the ABRE cis-element has sequence variability, and GCGT or even AAGT functions as an ABRE cis-element. Therefore, we examined the possibility that the sequence from −1087 to −1083 bp is an ABRE cis-element. However, a replacement of the sequence from AAGT to ACGT, a typical ABRE core sequence, was showed the significant reduction in luciferase activity. Taking into consideration of the replacement of the sequence (GAAGT; −1087 to −1083 bp) with its complementary sequence (CTTCA), it seems that the sequence dose not acts as an ABRE cis-element.
Fig. 1. Promoter analysis of the acerola GMP (MgGMP) gene.
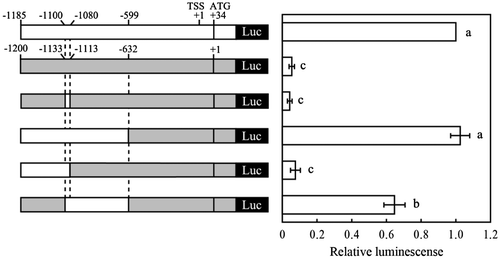
We also examined the contribution of the sequence (GAAGT) from −1087 to −1083 bp to MgGMP gene expression. Based on the position from initiation codons of MgGMP and AtGMP genes, the 5′-upstream region of MgGMP was replaced to that of AtGMP. Replacement of the sequence between −1133 and −1114 bp of the AtGMP promoter with the position corresponding sequence of the MgGMP promoter did not result in any significant increase in luciferase activity (Fig. ). An increase in luciferase activity was observed following replacement of the sequence from −1200 to −633 bp of the AtGMP promoter with the position corresponding sequence of the MgGMP promoter. In further replacement analysis, although the replacement of the sequence from −1200 to −1114 bp of the AtGMP promoter with the position corresponding sequence of the MgGMP promoter resulted in low luciferase activity, an increased luciferase activity was detected with the replacement of the sequence from −1133 to −633 bp of the AtGMP promoter with the position corresponding sequence of the MgGMP promoter. The results showed that the −1100 to −600 bp sequence of the MgGMP promoter is necessary for high MgGMP promoter activity, suggesting the presence of unknown cis-element(s), which function as enhancers of MgGMP expression, present in the −1080 to −600 bp sequence of the MgGMP promoter. However, because there is low sequence similarity between 5′-upstream regions of MgGMP and AtGMP, it is difficult to show putative cis-element(s) to enhance MgGMP expression at present study.
Fig. 2. Replacement analysis of Arabidopsis GMP promoter to acerola GMP promoter.
In this study, we found a new cis-element in the −1087 to −1083 bp sequence of the MgGMP promoter. However, so far as we know, there was no candidate cis-element for this sequence. In addition, MgGMP expression appears to be regulated by multiple cis-elements. In particular, the sequence including the cis-element found in this study, in the −1080 to −600 bp of the MgGMP promoter, is responsible for high MgGMP gene expression. In acerola,Citation9) the gene expression pattern of enzymes involved in AsA biosynthesis were similar between the fruits and leaves. In addition, MgGMP is highly expressed in leaves as well as fruits. However, it remains unclear whether the putative cis-element(s) found in this study function in acerola fruits. To further clarify data here, it will be necessary to investigate the transcriptional mechanism of MgGMP using acerola plants.
Author contribution
T. Kondo designed and performed the experiments, analyzed data, and wrote the paper. Y. Fujikawa gave the advice of research and paper. M. Esaka designed research and gave the advice of research and paper.
Funding
This work was supported by the Grant-in-Aid for Scientific Research (C) of JSPS KAKENHI [grant number 15K07394].
Disclosure statement
No potential conflict of interest was reported by the authors.
Acknowledgments
The sequence analysis was carried out at the Analysis Center of Life Science, Natural Science Center for Basic Research and Development, Hiroshima University.
Notes
Abbreviations: AsA, ascorbic acid; GMP, GDP-d-mannose pyrophosphorylase; ABRE, abscisic acid response element; RLU, relative light units; TSS, transcription start site.
References
- Noctor G, Foyer CH. Ascorbate and glutathione: keeping active oxygen under control. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:249–279.10.1146/annurev.arplant.49.1.249
- Barth C, Tullio MD, Conklin PL. The role of ascorbic acid in the control of flowering time and the onset of senescence. J Exp Bot. 2006;57:1657–1665.10.1093/jxb/erj198
- Mano J, Hideg E, Asada K. Ascorbate in thylakoid lumen functions as an alternative electron donor to photosystem II and photosystem I. Arch Biochem Biophys. 2009;429:71–80.
- Kotchoni SO, Jimenez-Lopez JC, Gao D, et al. Modeling-dependent protein characterization of the rice aldehyde dehydrogenase (ALDH) superfamily reveals distinct functional and structural features. PLoS ONE. 2009;5(7):e11516.
- Wheeler GL, Jones MA, Smirnoff N. The biosynthetic pathway of vitamin C in higher plants. Nature. 1998;393:365–369.
- Conklin PL, Norris SR, Wheeler GL, et al. Genetic evidence for the role of GDP-mannose in plant ascorbic acid (vitamin C) biosynthesis. Proc Nat Acad Sci USA. 1999;96(7):4198–4203.10.1073/pnas.96.7.4198
- Wang HS, Yu C, Zhu AJ, et al. Overexpression in tobacco of a tomato GMPase gene improves tolerance to both low and high temperature stress by enhancing antioxidation capacity. Plant Cell Rep. 2011;30:1029–1040.10.1007/s00299-011-1009-y
- Zhang Z, Wang J, Zhang R, et al. The ethylene response factor AtERF98 enhances tolerance to salt through the transcriptional activation of ascorbic acid synthesis in Arabidopsis. Plant J. 2012;71:273–287.10.1111/tpj.2012.71.issue-2
- Badejo AA, Fujikawa Y, Esaka M. Gene expression of ascorbic acid biosynthesis related enzymes of the Smirnoff–Wheeler pathway in acerola (Malpighia glabra). J Plant Physiol. 2009;166:652–660.10.1016/j.jplph.2008.09.004
- Badejo AA, Fujikawa Y, Esaka M. Analysis of GDP-d-mannose pyrophosphorylase gene promoter from acerola (Malpighia glabra) and increase in ascorbate content of transgenic tobacco expressing the acerola gene. Plant Cell Physiol. 2008;49(1):126–132.10.1093/pcp/pcm164
- Suekawa M, Fujikawa Y, Inada S, et al. Gene expression and promoter analysis of a novel tomato aldo-keto reductase in response to environmental stresses. J Plant Physiol. 2016;200:35–44.10.1016/j.jplph.2016.05.015
- Fujikawa Y, Nakanishi T, Kawakami H, et al. Split luciferase complementation assay to detect regulated protein–protein interactions in rice protoplasts in a large-scale format. Rice. 2014;7(1):11.10.1186/s12284-014-0011-8
- Kato N, Fujikawa Y, Fuselier T, et al. Luminescence detection of SNARE–SNARE interaction in Arabidopsis protoplasts. Plant Mol Biol. 2010;72(4–5):433–444.10.1007/s11103-009-9581-z
- Higo K, Ugawa Y, Iwamoto M, et al. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 1999;27(1):297–300.10.1093/nar/27.1.297
- Abe H, Urao T, Ito T, et al. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell. 2003;15:63–78.10.1105/tpc.006130
- Sarkar AK, Lahiri A. Specificity determinants for the abscisic acid response element. FEBS Open Bio. 2013;3:101–105.10.1016/j.fob.2013.01.006
