Abstract
A toxin-antitoxin system, vp1842/vp1843, locates within a superintegron on the Vibrio parahaemolyticus genome chromosome I whose toxin gene vp1843 encodes a DNA nicking endonuclease. We found that the vp1843 expression in Escherichia coli cells strongly induced chromosomal DNA degradation. On the basis of these observations, we discuss a possible physiological role of vp1842/vp1843 in V. parahaemolyticus.
Toxin-antitoxin (TA) systems are small genetic elements composed of two genes organized on an operon which encodes a stable toxin and a labile cognate antitoxin, respectively.Citation1,2) They were initially identified on low copy number plasmids, where they help to maintain plasmid stability by a mechanism known as post-segregational killing.Citation3,4) When a plasmid bearing a TA system is inherited, antitoxins neutralize the effects of toxins by direct protein–protein interactions. If the plasmid is not transmitted to a daughter cell, the unstable antitoxin is degraded while the stable toxin remains and acts on its cellular targets to kill the plasmid-free cells by interfering with vital cellular functions, including DNA replication and protein synthesis. Meanwhile, TA systems were found to be widely distributed and highly abundant in the genomes of bacteria and archaea, and they are proposed to function as metabolic stress response elements to manage their metabolism to cope with different sources of stress, although their biological functions are still under debate.Citation5)
In a previous study,Citation6) we found that gene clusters, vp1829/vp1830 and vp1842/vp1843, in the V. parahaemolyticus genome database have sequence homology to that encoding the Escherichia coli TA protein, DinJ/YafQ. Expression of the putative toxin gene, vp1843, in E. coli strongly inhibited cell growth, while co-production of the putative antitoxin gene, vp1842, neutralized this effect. In contrast, the expressions of vp1830 in the presence of 0.2% arabinose had no influence on cell growth. These results suggested that vp1842/vp1843 serve as a TA system in V. parahaemolyticus. We further found recently that Vp1843, unlike the E. coli homologue YafQ, has neither protein synthesis inhibitory activity nor ribonuclease activity, rather it has DNA nicking endonuclease activity.Citation7)
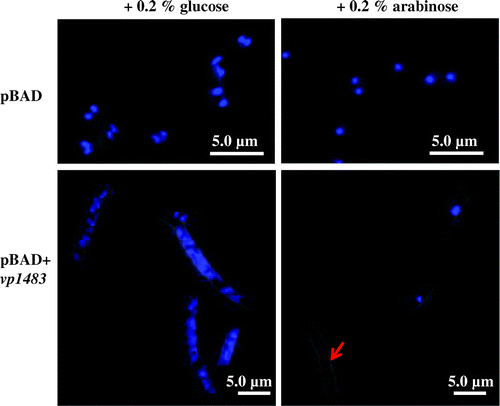
To gain insight into a physiological role of vp1842/vp1843 in V. parahaemolyticus, construction of a deletion of the vp1842 gene and an ectopic expression of the vp1843 gene in V. parahaemolyticus were carried out. However, these attempts were unsuccessful, suggesting extreme toxicity of vp1843 expression in the cells. Our previous study showed that the expression of vp1843 in E. coli cells caused morphological changes in the cells.Citation6) To gain more information about the physiological function of vp1842/vp1843, the E. coli cells (E. coli LMG194) expressing vp1843 were characterized using fluorescent microscopy. A single colony harboring pBAD/Myc-HisA-vp1843 was pre-cultured at 37 °C in RM medium containing 0.2% D-glucose (RMG) until an OD590 of 0.5. Then, the culture was divided into two equal parts; half of the cell was collected and resuspended in fresh RMG, while the other half was resuspended in fresh RMA (RM medium with 0.2% L-arabinose). Cells were exponentially grown in RMA medium for 3 h, washed in 0.9% NaCl, spotted on poly-L-lysine-coated slide glass and fixed in methanol. Nuclei were visualized with 1 μg/mL DAPI (4′,6-Diamidino-2-Phenylindole, Roche). Interestingly, a leak expression level of vp1843 (cultured in RMG medium) resulted in a filamentous cells (Fig. , left). Upon induction of vp1843, chromosomal DNA experienced a fast and extreme degradation, particularly as indicated by a red arrow (Fig. , right). This finding suggested that vp1843 expression results in the chromosomal DNA degradation.
Fig. 1. Microscopic analysis of E. coli cells repressing or expressing vp1843.
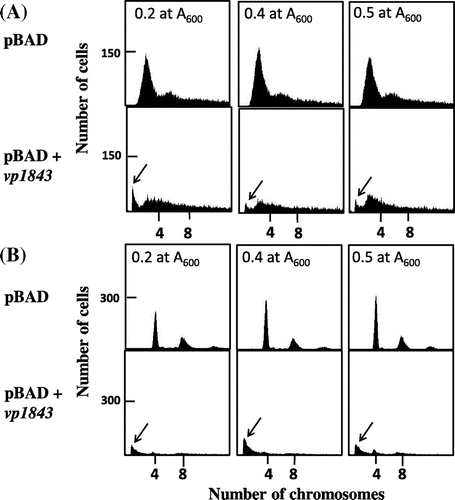
To examine the chromosomal degradation further, we analyzed the content of the E. coli chromosomal DNA in the presence or absence of the vp1843 gene in the pBAD vector using flow cytometry. E. coli cells containing pBAD/Myc-HisA or pBAD/Myc-HisA with vp1843 were incubated at 1 × 105 cells/mL in RMG medium containing 0.2% glucose and incubated at 37 °C, and when the culture reached 1 × 107 cells/mL, the exponentially growing culture in a phase of balanced growth was used for further analyses. Measuring the copy number of the chromosomal DNA was analyzed by the run-off replication method according to Adachi et al.Citation8) As shown in Fig. (A), the E. coli cells containing only the vector plasmid had the E. coli chromosomal DNA, while those harboring pBAD/Myc-HisA containing vp1843 had a smaller amount of chromosomal DNA and an additional peak at a position showing a smaller size of chromosomal DNA. This result suggested that a leak expression level of vp1843 (cultured in RMG medium in the presence of glucose) caused the chromosomal DNA degradation. To corroborate this assumption, the contents of the chromosomal DNA in the E. coli cells harboring only the vector plasmid or the plasmid with vp1843 were compared after 4 h incubation in the presence of rifampicin and cephalexin (Fig. (B)). As cephalexin inhibits cell division and rifampicin inhibits replication initiation, but not progress of replisomes, incubation with these drugs allows individual cells to complete the whole chromosome replication. As expected, the E. coli cells containing only the vector plasmid contained 4 and 8 copies of the chromosomal DNAs, whereas those harboring the plasmid with vp1843 had little chromosomal DNA and again an additional peak showing a smaller size of DNA (Fig. (B)). This result strongly suggested that the leak expression of vp1843 induced DNA degradation all over the E. coli chromosome.
Fig. 2. Flow cytometry of E. coli cells containing pBAD/Myc-HisA or pBAD/Myc-HisA with vp1843.
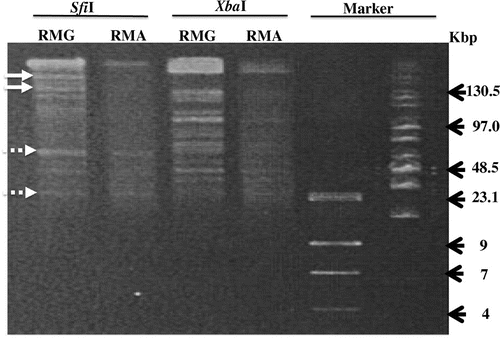
To corroborate the chromosomal degradation, the chromosome DNA was extracted and evaluated by pulsed-field gel electrophoresis (Fig. ).Citation9) Apparently, the E. coli cells grown in RMA had a smaller amount of chromosomal DNA than those grown in RMG. In addition, the restriction patterns of the chromosomal DNA isolated from E. coli cells grown in RMG were slightly distinct from those of the E. coli cells grown in RMA, as indicated in Fig. . This result suggested further that the vp1843 expression caused DNA degradation throughout the E.coli chromosome.
Fig. 3. Pulsed-field gel electrophoresis.
In bacteria, SOS is a global response to DNA damage, mediated by the recA-lexA genes, resulting in DNA repair and thereby in the survival of the bacterial culture. However, it has also been described that under severe DNA damage strong activation of RecA occurs, leading to the apoptosis-like death pathway, characterizing DNA fragmentation.Citation10) Recently, we found that Vp1843 has DNA nicking endonuclease activity.Citation7) It is thus likely that the vp1843 expression nicks the E. coli chromosomal DNA, which results in severe DNA damage, leading to DNA degradation and the apoptosis-like death. Interestingly, vp1842/vp1843 locates in a superintegron in the V. parahaemolyticus chromosome I.Citation11) The superintegrons, which are genomic islands at least 100 kb in size and contain a large number of open reading frames encoding proteins with no known function, have been discovered in the genomes of diverse proteobacterial species.Citation12) They are thought to be massive ancestral versions of multiresistance integrons, in which several antibiotic-resistant genes are located. It has been reported that when located in genomic islands, TA systems participate in stabilization, which is reminiscent of post-segregational killing mediated by plasmid-encoded TA systems.Citation13) Probably, when the superintegron including vp1842/vp1843 within the chromosome I is lost or damaged, the activated Vp1843 nicks the chromosomal DNA, which results in severe DNA damage, leading to DNA degradation and the apoptosis-like death. This assumption is consistent with the fact that construction of mutant strains, in which vp1842/vp1843 in the V. parahaemolyticus genome were knocked out by homologous recombination, was unsuccessful (Zhang et al. unpublished results). Thus, vp1842/vp1843 may be involved in the maintenance of the superintegron in the V. parahaemolyticus chromosome I in a manner similar to post-segregational killing.
Author contributions
J.Z. T.K. and M.K. conceived and designed the study and discussed the results. J.Z. S.T. and K.I. performed the experiments, and analyzed the data along with T.K. and M.K. M.K. wrote the manuscript in consultation with J.Z., H.I. M.H., and T.K.
Disclosure statement
No potential conflict of interest was reported by the authors.
Funding
This work was supported in part by a grant-in-aid for scientific research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan [grant number 26660090] to M.K. J. Zhang was sponsored by CSC (China Scholarship Council) [grant number 201308310430].
Acknowledgments
We are grateful to Prof. M. Ito and Associate Prof. N. Okino (Kyushu University) for their active interest and helpful discussions.
References
- Yamaguchi Y, Park JH, Inouye M. Toxin-antitoxin systems in bacteria and archaea. Annu Rev Genet. 2011;45:61–79.10.1146/annurev-genet-110410-132412
- Unterholzner SJ, Poppenberger B, Rozhon W. Toxin-antitoxin systems: biology, identification, and application. Mob Genet Elem. 2013;3(5):e26219.10.4161/mge.26219
- Ogura T, Hiraga S. Mini-F plasmid genes that couple host cell division to plasmid proliferation. Proc Natl Acad Sci USA. 1983;80:4784–4788.10.1073/pnas.80.15.4784
- Gerdes K, Rasmussen PB, Molin S. Unique type of plasmid maintenance function: postsegregational killing of plasmid-free cells. Proc Natl Acad Sci USA. 1986;83:3116–3120.10.1073/pnas.83.10.3116
- Page R, Peti W. Toxin-antitoxin systems in bacterial growth arrest and persistence. Nat Chem Biol. 2016;12:208–214.10.1038/nchembio.2044
- Hino M, Zhang J, Takagi H, et al. Characterization of putative toxin-antitoxin systems in Vibrio parahaemolyticus. J App Microbiol. 2014;117:185–195.10.1111/jam.2014.117.issue-1
- Zhang J, Ito H, Hino M, et al. A RelE/ParE superfamily toxin in Vibrio parahaemolyticus has DNA nicking endonuclease activity. Biochem Biophys Res Commun. 2017;489:29–34.10.1016/j.bbrc.2017.05.105
- Adachi S, Fukushima T, Hiraga S. Dynamic events of sister chromosomes in the cell cycle of Escherichia coli. Genes to Cells. 2008;13:181–197.10.1111/j.1365-2443.2007.01157.x
- Ribot EM, Fitzgerald C, Kubota K, et al. Rapid pulsed-field gel electrophoresis protocol for subtyping of Campylobacter jejuni. J Clin Microbiol. 2001;39:1889–1894.10.1128/JCM.39.5.1889-1894.2001
- Erental A, Kalderon Z, Saada A, et al. Apoptosis-like death, an extreme SOS response in Escherichia coli. mBio, 2014;5:e01426-14.
- Makino K, Oshima K, Kurokawa K, et al. Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V. cholera. The Lancet. 2003;361:743–749.10.1016/S0140-6736(03)12659-1
- Rowe-Magnus DA, Guerout AM, Biskri L. Comparative analysis of superintegrons: engineering extensive genetic diversity in the vibrionaceae. Genome Res. 2003;13:428–442.10.1101/gr.617103
- Hallez R, Geeraerts D, Sterckx Y. New toxins homologous to ParE belonging to three-component toxin-antitoxin systems in Escherichia coli O157:H7. Mol Microbiol. 2010;76:719–732.10.1111/mmi.2010.76.issue-3
