Abstract
The presence of d-aspartate (d-Asp), a biologically rare amino acid, was evaluated in 38 species of marine macroalgae (seaweeds). Despite the ubiquitous presence of free l-Asp, free d-Asp was detected in only 5 species belonging to the Sargassaceae family of class Phaeophyceae (brown algae) but not in any species of the phyla Chlorophyta (green algae) and Rhodophyta (red algae). All other members of Phaeophyceae, including 3 species classified into the section Teretia of Sargassaceae did not contain d-Asp. These results indicate that the presence of free d-Asp in marine macroalgae is restricted only to the Sargassaceae family, excluding the species in the section Teretia.
Presence of d-Aspartate in macroalgae is restricted to the Sargassaceae family except Teretia section
In the past four decades, various d-amino acids have been found in animals and plants [Citation1,2]. Within aquatic animals, d-aspartate (d-Asp) and d-alanine have been identified in cephalopods [Citation3,4], crustaceans [Citation5], and mollusks [Citation6]. Recently, it was reported that d-Asp and aspartate racemase [EC 5.1.1.13] are involved in anaerobic energy metabolism in the bivalve mollusk Anadara broughtonii (formerly Scapharca broughtonii) [Citation7,8]. Although little attention has been paid to the natural occurrence and physiological role of d-Asp in macroalgae, study on the presence of d-amino acids in macroalgae commenced following the observation of high levels of free d-Asp in the marine macroalga Sargassum fusiforme [Citation9]. S. fusiforme germinates in late summer and grows from autumn to spring in the sea near Iwate Prefecture, northern part of the largest island of Japan, showing rapid growth particularly in early spring. The d-Asp content also increases during this early growth period. These findings suggest that d-Asp plays an important role in the early growth stages of S. fusiforme [Citation9], but the exact biological role of d-Asp in this alga is yet to be clarified. To gain deeper insight into the biological role of d-Asp in S. fusiforme, we prepared a polyclonal antibody against d-Asp and confirmed that d-Asp is present in the intracellular structure of S. fusiforme [Citation10]. These results exclude the possibility that d-Asp is derived from attached or symbiotic organisms, such as marine bacteria.
Nagahisa et al. [Citation11] reported the presence of free d-Asp in some marine macroalgae in Japan. They found that d-Asp was present at high levels, with concentrations proportional to those of l-Asp, in Costaria costata, S. fusiforme, and S. yezoense that belong to Phaeophyceae (brown algae). However, d-Asp was not found in many other species of macroalgae, and even if it was detected, it was present only in trace amounts. S. fulvellum, S. fusiforme, and S. yezoense thrive in the same intertidal zones in the sea. Although S. fusiforme and S. yezoense are reported to have high levels of d-Asp, its content in S. fulvellum is very low. Thus, the question arises as to why the amount of d-Asp largely differs in marine macroalgae belonging to the same genus, Sargassum. In the present study, the distribution of d-Asp in many marine macroalgae, particularly those belonging to the family Sargassaceae, was examined to clarify the seaweeds that contain d-Asp.
This is the first report presenting a taxonomical view of the presence of free d-Asp by relating it to the classification of marine macroalgae.
Materials and methods
Materials
Marine macroalgae were collected from the coast of Iwate and Kanagawa Prefecture, Japan, in 2013 and 2014 as shown in Table . Algal extracts were prepared from 5 species of Chlorophyta (green algae), 15 species of Rhodophyta (red algae), and 18 species of Phaeophyceae. All chemicals, including d- and l-Asp, o-phthalaldehyde (OPA), and N-acetyl-l-cysteine (NAC), were purchased from Wako Pure Chemicals (Osaka, Japan), and were of analytical-grade purity. Milli-Q water for the experiments was obtained by purifying distilled water using a Simplicity UV (Merck Millipore, Billerica, MA, USA) system.
Table 1. Distribution of free D- and L-aspartate in marine macroalgae.
Preparation of algal extract
A part of the thalli of each sampled alga (about 10 g wet weight) was rinsed with distilled water to remove attached foreign materials, such as small organisms. The algae were dried by wrapping the thalli tightly with paper towel and a portion of the thallus was homogenized with 10 volumes of 80% (v/v) ethanol using a polytron homogenizer (PT-K, Kinematica, Lucerne, Switzerland) followed by centrifugation at 50,000 × g at 4 °C for 15 min. The resultant precipitate was extracted with the same volume of ethanol solution for maximal extraction. The supernatants (ethanolic extracts) were combined and evaporated to dryness under reduced pressure at 40 °C. The residue was dissolved in a small amount of Milli-Q water and then rinsed with a sufficient amount of diethyl ether to remove pigments and fatty materials. The aqueous solution layer was transferred to an eggplant-shaped flask and evaporated to dryness. The obtained residue was dissolved in 50 mL of Milli-Q water and stored at −20 °C until further analysis with high-performance liquid chromatography (HPLC).
Derivatization of amino acids with OPA/NAC
Each sample extract was passed through a 0.20 μm filter. For derivatization of amino acids, 10 μL of the sample solution was mixed with 70 μL of a saturated sodium borate solution and 20 μL of a mixture of 10 mg OPA and 10 mg NAC in 1 mL of methanol. After allowing the mixture to react for 1 min at room temperature, the reaction mixture was injected directly into the HPLC system.
Determination of d- and l-aspartate
Chromatography was performed as described by Nimura and Kinoshita [Citation12] with some alterations [Citation13]. Chromatographic analysis was performed with a JASCO (Tokyo, Japan) HPLC system consisting of a PU-2089 quaternary gradient pump with a degasser, a CO-2065 column oven, an AS-2057 autosampler with a cooling system, an FP-2020 fluorescence detector, and a ChromNAV data processor. The analytical column was a reversed-phase ODS-80TS (4.6 × 250 mm) (Tosoh, Tokyo, Japan) with a guard column (3.2 × 15 mm) packed with the same resin. Elution was performed with a mixture of solvent A (50 mm sodium acetate buffer at pH 5.6) and solvent B (methanol:solvent A = 80:20) at 40 °C; the flow rate was set at 1.0 mL・min−1. For fluorometric detection of eluted OPA/NAC derivatives, the excitation and emission wavelengths were set to 350 and 450 nm, respectively. The elution gradient was set as follows: 0–20 min, 0–20% solvent B in solvent A; 20–35 min, 20% solvent B in solvent A.
Results
Distribution of free d- and l-aspartate in marine macroalgae
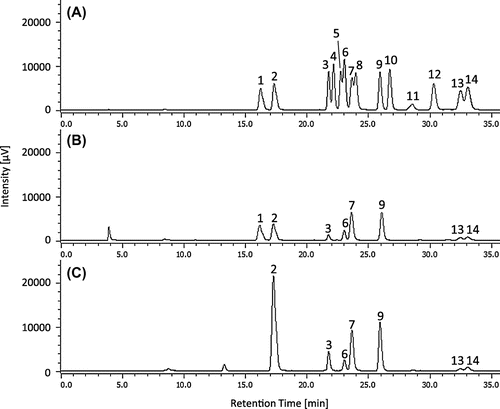
Typical HPLC chromatograms of the converted d- and l-enantiomers of standard amino acids, an extract of S. fusiforme with the presence of d-Asp, and an extract of S. thunbergii with no d-Asp are shown in Figure . Retention times of both d- and l-Asp derivatives fully corresponded with each peak obtained for extracts of S. fusiforme (Figure ; peak number 1 and 2).
Figure 1. Typical HPLC chromatograms of (A) the OPA/NAC derivatives of d- and l-enantiomers of standard amino acids, (B) an extract of the macroalgae Sargassum fusiforme and (C) Sargassum thunbergii.
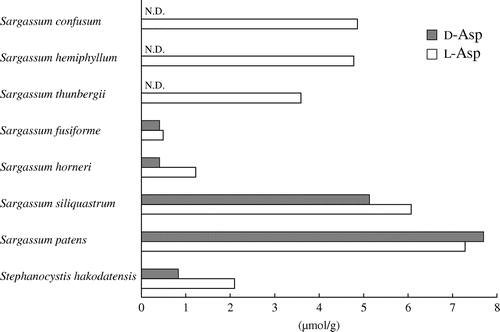
Although l-Asp was detected in all species examined, d-Asp was not detected in species belonging to Chlorophyta, Rhodophyta, and most species of Phaeophyceae. However, d-Asp was present in some species of Phaeophyceae belonging to the genus Sargassum and Stephanocystis of Sargassaceae (Table ).
Figure shows a concentration of d- and l-Asp in Sargassaceae. No traces of d-Asp were detected in S. confusum, S. hemiphyllum, and S. thunbergii. However, S. fusiforme, S. horneri, S. siliquastrum, and S. patens contained d-Asp in significant concentrations (0.41, 0.41, 5.13, and 7.70 μmol・g−1 wet weight, respectively). Stephanocystis hakodatensis also contained 0.83 μmol・g−1 wet weight of d-Asp. In addition, the percent content of [D/(D + L)] × 100 (%) was generally high in S. fusiforme, S. siliquastrum, and S. patens (45.6, 45.8, and 51.4 %, respectively), whereas that in S. horneri and S. hakodatensis was 25.2 and 28.4%, respectively.
Discussion
The presence of free d-Asp in marine macroalgae was confirmed using pre-column HPLC amino acid analysis. d-Asp was detected only in 5 distinguishable species out of the total of 38 species examined (13% of the species). The macroalgae containing d-Asp all belonged to the Sargassaceae family, indicating that free d-Asp is specifically presents in the family Sargassaceae of the class Phaeophyceae, although S. confusum, S. hemiphyllum, and S. thunbergii, which also belong to Sargassaceae, contained no d-Asp (Table ).
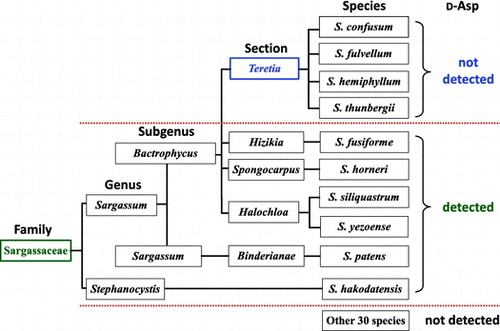
Generally, Sargassaceae exist at similar locations i.e. on rocks in the lower intertidal zone to the infralittoral zone; Their growth season is also recognized as the duration from autumn to spring in Japan. Therefore, we presumed that the presence of d-Asp is related to algae’s taxonomic ranks rather than their habitat. In biological classification, taxonomic ranks comprise species, genus, family, order, class, phylum (division), and kingdom. However, taxonomic ranks in botany include the secondary ranks, such as “section” between genus and species. Thus, the knowledge about detailed classification of Phaeophyceae should be considered to gain deeper insight into the diverse distribution of d-Asp in Sargassaceae. According to detailed taxonomic ranks, it was found that d-Asp was detected in the entire family Sargassaceae, except the section Teretia (Figure ). Therefore, the presence of free d-Asp in macroalgae is taxonomically restricted to certain sections in Sargassaceae.
Figure 3. Convincing interpretation for the presence of free d-aspartate in Sargassaceae. Infrageneric classification is based on the algae database.
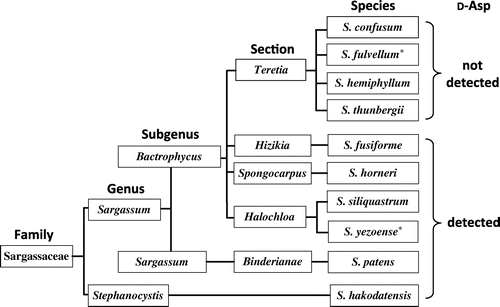
The taxonomical position of S. fusiforme has been discussed for a long time [Citation14]. At present, S. fusiforme belongs to the section Hizikia in the genus Sargassum [Citation14]. The genus of Sargassum in to subgenus Bactrophycus is mainly based on the morphology of the basal part supplemented by the morphology of receptacles. In the present study, we reported high accumulation of d-Asp in S. fusiforme from the section Hizikia, but its absence in species belonging to the section Teretia. Although the section Hizikia is positioned near the section Teretia morphologically, the presence of d-Asp is unique.
Nagahisa et al. [Citation11] reported a very low content of d-Asp in S. fulvellum from the section Teretia (0.0012 μmol・g−1 wet weight) compared to that in S. yezoense (0.0485 μmol・g−1 wet weight) from the section Halochloa, whereas C. costata contained 0.066 μmol・g−1 wet weight of d-Asp; these results were not corroborated in the present study (Table ). This difference in d-Asp content may have resulted from different protocols implemented during the preparation of the samples before analyses. Namely, in the present study, thalli were carefully cleaned from any attached organisms and the identification of macroalgae species was verified.
It is known that the content of amino acids in marine macroalgae changes with seasons and/or locations. d-Asp is present in S. fusiforme throughout the year although its level vary with time [Citation9]. Similarly, the presence of d-Asp in S. fusiforme was confirmed in thalli of samples collected at both Iwate and Kanagawa (data not shown). These results suggest that the presence of d-Asp in S. fusiforme is independent of season and/or location. It is speculated that the d-Asp is present in other marine macroalgae of the family Sargassaceae, except the section Teretia, regardless of the season and/or location, although more detailed studies are necessary.
The available information on the biological role and metabolic pathway of d-Asp in marine organisms is limited. It was reported that d-Asp and aspartate racemase are involved in anaerobic energy metabolism in the bivalve mollusk, A. broughtonii [Citation7,8]. In contrast, the biological role and metabolic pathway of d-Asp in marine macroalgae remain unclear. Funakoshi et al. [Citation15] reported the presence of d-amino acid transaminase [EC 2.6.1.21], which catalyzes transamination between d-Asp and other d-amino acids, in the terrestrial plant, Arabidopsis thaliana. Recently, Ito et al. [Citation16] reported that mammalian enzyme serine racemase, the primary enzyme responsible for brain d-serine production also catalyzes Asp racemization. Unfortunately, aspartate racemase, serine racemase, and/or d-amino acid transaminase activity remain to be detected in any macroalgae through biochemical analyses [Citation2]. Moreover, there is no genome information about species belonging to the Sargassaceae. Thus, it is necessary to identify and characterize enzymes involved in the metabolism of d-Asp in Sargassaceae through molecular and biochemical testing, which provides novel insights into lineage-specific presence, biosynthetic pathways, and physiological functions of d-Asp in marine brown macroalgae.
Author contribution
T. Yokoyama preformed the experimental design and experiments. Experimental interpretation of data was conducted by all authors. T. Yokoyama wrote the paper, and the other authors commented on the manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
Funding
This work was supported in part by JSPS KAKENHI [grant numbers 16K07852, 15H04539].
Acknowledgments
We would like to express our thanks to Dr. Nobuyoshi Nanba and Mr. Kousuke Takada of Kitasato University for their assistance in this study.
References
- Soda K. An essay on d-Amino acids: retrospection and perspective. In: Konno R, Brückner H, D’Aniello A, et al., editors. d-Amino acids: a new frontier in amino acid and protein research–practical methods and protocols. New York (NY): Nova Science; 2007. p. 3–14.
- Yokoyama T, Mikami K. d-Amino acids and amino acid racemases in seaweeds. In: Pomin VH, editor. Seaweeds: agricultural uses, biological and antioxidant agents. New York (NY): Nova Science; 2014. p. 135–155.
- D’Aniello A, Giuditta A. Identification of D-aspartic acid in the brain of octopus vulgaris lam. J Neurochem. 1977;29:1053–1057.10.1111/jnc.1977.29.issue-6
- D’Aniello A, Giuditta A. Presence of D-aspartate in squid axoplasm and in other regions of the cephalopod nervous system. J Neurochem. 1978;31:1107–1108.10.1111/jnc.1978.31.issue-4
- D’Aniello A, Giuditta A. Presence of d-alanine in crustacean muscle and hepatopancreas. Comp Biochem Physiol. 1980;66:319–322.
- Felbeck H, Wiley S. Free D-amino acids in the tissues of marine bivalves. Biol Bull. 1987;173:252–259.10.2307/1541877
- Shibata K, Watanabe T, Yoshikawa H, et al. Nucleotides modulate the activity of aspartate racemase of Scapharca broughtonii. Comp Biochem Physiol. 2003;134:713–719.10.1016/S1096-4959(03)00031-9
- Watanabe T, Shibata K, Kera Y, et al. Effects of hypoxic and osmotic stress on the free d-aspartate level in the muscle of blood shell Scapharca broughtonii. Amino Acids. 2005;28:291–296.10.1007/s00726-005-0188-7
- Nagahisa E, Kanno N, Sato M, et al. Variations in d-aspartate content with season and part of Hizikia fusiformis. Fish Sci. 1994;60:777–779.10.2331/fishsci.60.777
- Yokoyama T, Amano M, Sekine M, et al., Immunohistochemical localization of endogenous d-aspartate in the marine brown alga sargassum fusiforme. Biosci Biotechnol Biochem. 2011;75:1481–1484.10.1271/bbb.110184
- Nagahisa E, Kanno N, Sato M, et al. Occurrence of free d-aspartic acid in marine macroalgae. Biochem Int. 1992;28:11–19.
- Nimura N, Kinoshita T. o-Phthalaldehyde–N-acetyl-l-cysteine as a chiral derivatization reagent for liquid chromatographic optical resolution of amino acid enantiomers and its application to conventional amino acid analysis. J Chromatogr. 1986;352:169–177.10.1016/S0021-9673(01)83377-X
- Yokoyama T, Kan-no N, Ogata T, et al. Presence of free d-amino acids in microalgae. Biosci Biotechnol Biochem. 2003;67:388–392.10.1271/bbb.67.388
- Stiger V, Horiguchi T, Yoshida T, et al. Phylogenetic relationships within the genus Sargassum (Fucales, Phaeophyceae), inferred from ITS-2 nrDNA, with an emphasis on the taxonomic subdivision of the genus. Phycol Res. 2003;51:1–10.10.1111/pre.2003.51.issue-1
- Funakoshi M, Sekine M, Katane M, et al. Cloning and functional characterization of Arabidopsis thaliana d-amino acid aminotransferase – d-aspartate behavior during germination. FEBS J. 2008;275:1188–1200.10.1111/ejb.2008.275.issue-6
- Ito T, Hayashida M, Kobayashi S, et al. Serine rasemase is involved in d-aspartate biosynthesis. J Biochem. 2016;160:345–353.10.1093/jb/mvw043