ABSTRACT
Considering that fish grows in a complex aquatic environment, there is an increasing interest in fish β-defensins, which is an important group of antimicrobial peptides (AMPs). In this study, grass carp (Ctenopharyngodon idella) β-defensin 1 (gcdefb1) was isolated using homology cloning technology. Tissue distribution assay showed that gcdefb1 transcripts were expressed with the highest levels in brain and liver, followed by some mucous tissues. To examine gcDefb1 bioactivities, the recombinant gcDefb1 proteins fused with thioredoxin tag protein (Trx) (Trx-Defb1) were induced for production in Escherichia coli Rosetta-gami2(DE3)pLysS under optimal expression conditions. The antibacterial activity of Trx-Defb1 against Aeromonas hydrophila was assessed and its minimum inhibitory concentration (MIC) was 36 μM. Interestingly, Trx-Defb1 significantly inhibited LPS-induced Tnf-α (gcTnf-α) secretion and nitric oxide production in grass carp head kidney monocytes/macrophages (HKM), although Trx-Defb1 alone had no effect. Our studies provide the first evidence of fish β-defensin 1 engaging in both antimicrobial and inflammation suppression process.
Graphical Abstract
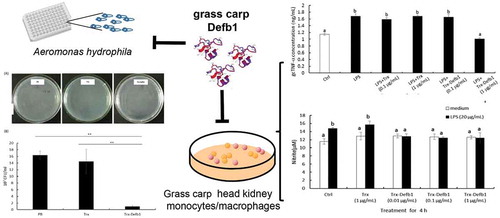
Dual role of grass carp Defb1 in inflammation control via antimicrobial activity against pathogen and immuno-suppressive modulation for host.
Defensins are a family of small, cysteine-rich and amphipathic AMPs that are widely distributed in plants, animals and fungi. Defensins can be classified into three categories, α-, β- and θ-defensins based on their disulphide bonding pattens. So far, β-defensin is the only type of category for fish. Various fish β-defensins widely distribute over spatial and temporal spans in epithelial cells and leukocytes of mucous membranes of the respiratory, genitourinary and gastrointestinal tracts. In these cells and tissues, they constitute the first line of defense against microbial infections, and their expression can be upregulated easily by pathogen-associated molecular patterns (PAMPs).
Despite of their conserved 6-cysteine motif and common tertiary structure, the amino acid sequences of β-defensins diversify considerably, either across the different species or within the same species. The sequence diversity of fish β-defensins contributes to their species-dependent broad spectrum of antimicrobial activity, which have been demonstrated in a few species, such as medaka [Citation1], channel catfish [Citation2], nile tilapia [Citation3], and atlantic cod [Citation4].
Besides microbicidal activities, mammalian AMPs show immune-modulatory activities. In the most cases, mammal AMPs harbor some abilities related to inducing inflammation, including promoting chemotactic effect, activating antigen presenting cells and cytokine induction [Citation5–Citation7]. In contrast, low levels of human β-defensin (hDEFB) may maintain a noninflammatory environment by neutralizing the effects of commensal and pathogenic microbial antigens [Citation8]. However, few reports has demonstrated the role of fish β-defensins on immune modulation.
Grass carp is largely farmed fish with huge economical importance and Aeromonas hydrophila is well known as one of the major causative agents of carp diseases [Citation9,Citation10]. In the present study,we isolated gcdefb cDNA using homology cloning technology. Sequence alignment and phylogenetic analysis indicated that this molecule belonged to the β-defensin 1 group of fish. Tissue distribution assay revealed that gcdefb1 transcripts displayed constitutive expression patterns with the highest expression in brain and liver. Subsequently, we prepared the recombinant protein of gcDefb1 with Trx fusion partner (Trx-Defb1). Trx-Defb1 showed antibacterial activity against Aeromonas hydrophila in intermediate level. Importantly, further study displayed Trx-Defb1 was able to suppress LPS-induced gcTnf-α secretion and nitric oxide production in grass carp HKM. In all, this is the first time clarifying the dual role of fish β-defensin 1 in inflammation control via antimicrobial activity against pathogen and immuno-suppressive modulation for host.
Materials and methods
Fish rearing
One-year-old Chinese grass carp with body weight of about 750 g were purchased from Chengdu Tongwei Aquatic Science and Technology Company (Chengdu, China). Fish were acclimated to the aquarium for two weeks before use. All experiments complied with the Regulation of Animal Use in Sichuan Province, China.
Molecular cloning of gcdefb1
Total RNA was extracted from liver with Tripure Isolation Reagent (Roche, Basel, Switzerland). RNA concentration was measured by spectrophotometry and integrity was determined by electrophoresis analysis. About 5 μg total RNA was reversely transcribed to cDNA using M-MLV Reverse Transcriptase (Promega, Madison, WI, USA). The degenerate primers of gcdefb1 pF and pR () were designed based on the conserved regions of β-defensin sequences of other fish, and used for the partial cDNA sequence of gcdefb. The full-length sequence of gcdefb1 was pulled out using 3ʹ and 5ʹ RACE System for Rapid Amplification of cDNA Ends (QIAGEN, Valencia, CA, USA) with the gene specific primers ().
Table 1. The primers used in the present study.
Sequence and phylogenetic analysis
The potential signal peptide of deduced amino acid sequence of gcDefb1 was predicted by SignalP 4.1 program (http://www.cbs.dtu.dk/services/SignalP/). The molecular weight (MW), the isoelectric point (pI) and the net charge of the peptide were calculated using the Antibacterial Peptides Database (APD) [Citation11]. The multiple alignments were made using DNAMAN software (Lynnon Biosoft, Pointe-Claire, Canada). Phylogenetic tree was constructed using MEGA7.1 software [Citation12] based on Neighbor-Joining method [Citation13] with the bootstrapping of 1000 repetitions.
Real-time quantitative PCR
The gene-specific intron-spanning primers () were designed for real-time quantitative PCR (qPCR). The qPCR was performed at a final volume of 10 μL containing 5 μL of RealMasterMix (Tiangen, China) and tissue cDNA as templates. The reactions were done on CFX96TM Real-time detection system (Bio-Rad, Hercules, CA, USA) for 35 cycles with 20 s at 94℃ for denaturation, 20 s at 54℃ for annealing, 20 s at 68℃ for extension. The standard curve was generated with 10-fold serial dilutions (from 10−1 to 10−6 fmol/μL) of the plasmid containing gcdefb1. Parallel measurement of β-actin was conducted to serve as an internal control.
Construction, expression and purification of Trx-Defb1
The DNA fragment coding gcDefb1 mature peptide (25–67 a.a.) was amplified by phusion high fidelity DNA polymerase (Finnzymes, Espoo, Finland) using primers rF and rR (). The fragment was subcloned into pGEM-T easy (Promega, Madison, WI, USA) and then ligated into pET-32a(+) between BamHI and XhoI to obtain the recombinant plasmid of pET-32a(+)/gcDefb1. The positive clone was confirmed by sequencing and transformed into E. coli Rosetta-gami2(DE3)pLysS (Novagen, Madison, WI, USA) for expression. The expression and purification procedure followed the pETman mannual (Novagen, San Diego, CA) with a brief modification. The induction was initiated by 1 mM isopropyl-β-D-thiogalactoside (IPTG, Merck, Darmstadt, Germany) at 30°C for 4 h until the growth to an optical density at 600 (OD600) of 1.0. In parallel, pET-32a(+) vector was transformed into Rosetta-gami2(DE3)pLysS to obtain Trx which served as a negative control.
Antibacterial assay of Trx-Defb1
A pathogenic strain of Aeromonas hydrophila subsp. hydrophila (A. hydrophila, ATCC 7966) was supplied by State Collection Center of Aquatic Pathogen, Shanghai Ocean University (Shanghai, China). Trx-Defb1 (1.6 μg/μL) were gradually diluted in 2-fold with phosphage buffer (PB,10mM, pH 7.0) to a series of dilutions. A. hydrophila was cultured to OD600 ≈ 0.4 and was adjusted into 1 × 104 colony forming units (CFU)/mL before use. Each dilution of Trx-Defb1 (100 μL) was mixed with 100 μL of bacterial inoculum in 96-well microtiter plate and incubated at 28°C overnight. MIC was defined as the minimum concentration of Trx-Defb1 used at which no visible turbidity of bacterial inoculum was observed at OD600. Additionally, the antibacterial activity of Trx-Defb1 was further confirmed by liquid bacteria inhibitory assays against A. hydrophila reported in literatures [Citation14,Citation15]. In brief, 100 μL of the bacterial liquid was inoculated into an aseptic 10-mL glass tube containing 400 μL of the drug (1 μg/μL) and incubated at 28°C. PB (10 mM, pH 7.0) only or Trx solution (1 μg/μL) were used as the controls. After overnight incubation, bacterial culture was diluted and spread on plates to count CFU/mL.
Isolation cells
Grass carp HKM and peripheral blood lymphocytes (PBLs) were prepared by discontinuous density gradient centrifugation as previously described with a little modification [Citation16]. Cells were seeded at a density of 9 × 105/well in 48-well plate (BD Biosciences, San Jose, USA) and incubated at 28°C under 5% CO2 and saturated humidity overnight. And then, the non-adherent cells were removed by washing twice and the adherent cells were used for drug treatment.
ELISA of gcTnf-α secretion
Competitive-inhibition ELISA was conducted to measure the concentrations of gcTnf-α secreted into the culture medium of grass carp HKM. The details of the method and the preparation of gcTnf-α polyclonal antibody (gcTnf-α Ab) were described previously [Citation17].
Determination of nitric oxide (NO) production
Grass carp HKM were treated with Trx-Defb1 (0.01–1 μg/mL), LPS (20 μg/mL) and Trx (1 μg/mL) or the combination of the drugs as indicated and the supernatant was collected for NO assay. After treatment, NO assay was carried out using the Griess reaction [Citation18]. All drug treatments were performed in quadruplicate and each experiment was independently repeated for three times.
Statistical analysis
Each experiment was repeated at least three times. Data were presented as mean ± SEM of four individual samples (N = 4) in each experiment. Statistical analysis was conducted by one-way analysis of variance (ANOVA) followed by Tukey’s test using the Statistical Product and Service Solutions 19.0 (SPSS, Chicago, USA). Student’s t-test was performed for comparing two groups. Differences were considered statistically significant at p<0.05.
Results and discussion
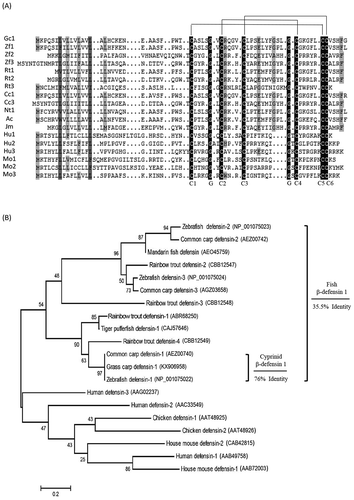
In the present study, gcdefb1 was isolated and cloned (GenBank Numbers: MG652749). The multiple alignment showed that in common with β-defensins of other species [Citation5], gcdefb1 contained eight highly conserved amino acid residues, including six C-terminal cysteines and two glycines separately ()). The phylogenetic tree showed that gcdefb1 were clustered with its homologs of cyprinid species with 76% identity. However, the amino acid sequences of fish β-defensins are highly variable, for the average identity being below 35.5% ()). The low homology of fish β-defensins indicates that they might possess different molecular characteristics.
Figure 1. Multiple alignment and phylogenetic tree analysis.
(A) Multiple alignment of the deduced amino acid of β-defensins from grass carp and other species. Black shade indicates identical amino acids and gray shade indicates similar amino acid. C1-C6 and G represent six conservative cysteines and two glycine residues, respectively. (B) Phylogenetic tree of β-defensins. Values at the forks indicate the percentage of trees in which this grouping occurred after bootstrapping (1000 replicates). Scale bar shows number of substitutions per base. Accession numbers of the used sequences are shown in brackets. The identity of amino acid sequence of cyprinid (76%) or fish species (35.5%) is shown.
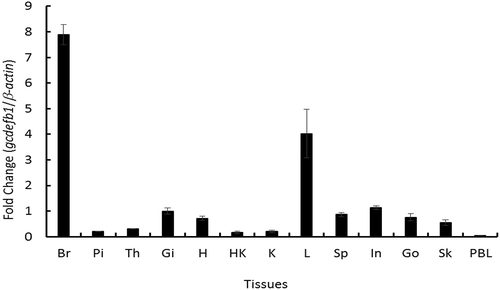
In agreement with this, fish β-defensin gene expression profiles appear species-dependent [Citation19]. For example, common carp β-defensin 1 is only detected in skin, not in liver, brain or gill [Citation19], and rainbow trout β-defensin 1 transcripts are only found in skin, too [Citation20]. Some other β-defensins, like hDEFB1 and zfDefb-1 [Citation21,Citation22], showed ubiquitous expression. In this study, gcdefb1 transcripts were constitutively expressed in all examined tissues with the highest levels in brain and liver, to the lesser extent in intestine, spleen, gill, gonad, heart and skin, and with the low levels in pituitary, thymus, head kidney, kidney and PBLs (). In line, gcdefb1 transcripts were detected in several mucous tissues including gill, skin and intestine. However, the highest expression of gcdefb1 was found in brain but not in mucosal tissues. The role of gcdefb1 in fish brain is unknown although a recent study reports the ability of AMPs against microbes in human brain [Citation23]. The species-dependent expression patterns indicate possible functional discrepancy of β-defense 1 in different fish species, promoting us to examine the bioactivity of gcDefb1.
Figure 2. Tissue distribution of gcdefb1 transcripts in grass carp.
Total RNA was extracted from tissues of 5 fish (N = 5) including brain (Br), pituitary (Pi), thymus (Th), gill (Gi), heart (H), head kidney (HK), kidney (K), liver (L), spleen (Sp), intestine (In), gonad (Go), Skin (Sk), and PBLs. The levels of gcdefb1 transcripts were measured by qPCR. The relative levels of gcdefb1 were normalized by β-actin level in the same sample and were presented as the fold changes of that in gill.
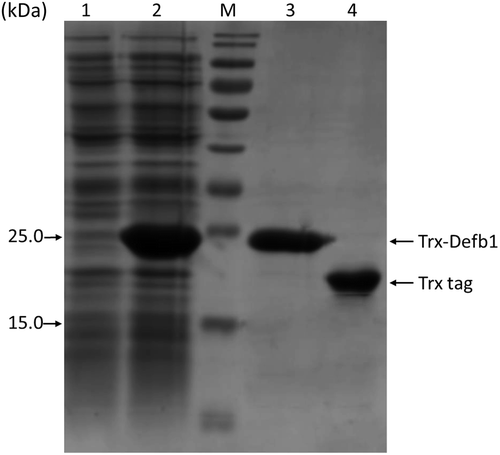
In order to study the function of gcdefb1, we prepared Trx-Defb1. Trx helps disulfide bond formation, making it a candidate for fusion expression. Considering that Trx-Defb1 might inhibit cell growth, the growth phase was prolonged by initiating the induction when OD600 reached 1.0, higher than 0.6–0.8 which most protocols recommended. Meanwhile, plasmid pLysS in Rosetta-gami2(DE3)pLysS host strain aids to stabilize plasmids containing toxic inserts and inhibit the basal expression level initiated by T7 RNA polymerase even before induction. Thus, Rosetta-gami2(DE3)pLysS harboring pET-32a(+)/gcDefb1 was induced for a short period (4 h). SDS-PAGE analysis showed that almost all the untargeted protein flowed through (Lane 1 in ) or was washed off by 80 mM imidazole off the column. The purity of Trx-Defb1 (22 kDa) reached more than 95% (Lane 3 in ). In parallel, Trx (20.3 kDa, Lane 4 in ) was obtained to be used as control.
Figure 3. SDS-PAGE analysis of Trx-Defb1 purity.
Lane 1 and Lane 2, the flow-through sample and sample before purification; M, molecular weight marker (kDa); Lane 3, purified Trx-Defb1 fusion protein; Lane 4, purified Trx protein.
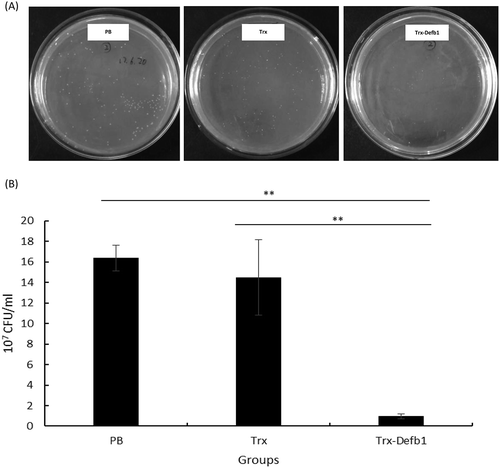
As a critical characteristic of AMPs, antibacterial activity has been evaluated in several fish β-defensins. MIC assay revealed that Trx-Defb1 inhibited the growth of A. hydrophila at a concentration of 36 μM. To exclude the possibility of contamination, the liquid bacteria inhibitory assay was performed. ) showed the representative plates of PB-, Trx- and Trx-Defb1-treated bacteria inoculum. The colonies on plates were round, smooth, semitransparent and homogenous, demonstrating that no contamination occurred. The bacterial growth in Trx-Defb1-treated group was sharply inhibited by more than 90% compared with PB and Trx groups ()). Thus, gcDefb1 possessed the moderate antibacterial activity against A. hydrophila compared with medaka (about 3 μM) [Citation1] and nile tilapia (about 100 μM) [Citation3]. This is in accordance with the property of gcDefb1. In fact, the net charge of gcDefb1 is as low as 0.61 at pH 7.4 compared with the average net charge (+4.18) of all β-defensins collected in APD [Citation11]. Furthermore, Trx-Defb1 showed the ability to inhibit the growth of A. hydrophila compared with Trx and PB, indicating that Trx as fusion partner did not affect the bioactivity of Defb1.
Figure 4. Antimicrobial activities of Trx-Defb1 against A. hydrophila.
(A) The representative plate cultures in the 10−5 dilution of PB, Trx (1 μg/μL) and Trx-Defb1 (1 μg/μL) co-incubated bacterial inoculum. (B) The statistics of CFU/mL in three groups indicated above. The “**” denotes the significant differences at p < 0.05 between Trx-Defb1 group versus PB or Trx group.
In respect to the modulation of immune response, human defensins have both pro- and anti- inflammatory effects in the immune system. However, the action of fish β-defensin in immuno-modulation remains elusive. gcTnf-α is an early responsive proinflammatory gene to antigen stimulation and promotes the induction of the other inflammatory cytokines in both physiological and pathological processes [Citation24]. Additionally, nitric oxide production can also defense against pathogens, while excessive amount will cause tissue or DNA damage and lead to chronic inflammation [Citation25]. Thus, it is widely accepted that TNF-α and nitric oxide production are the classical parameters of inflammation occurring.
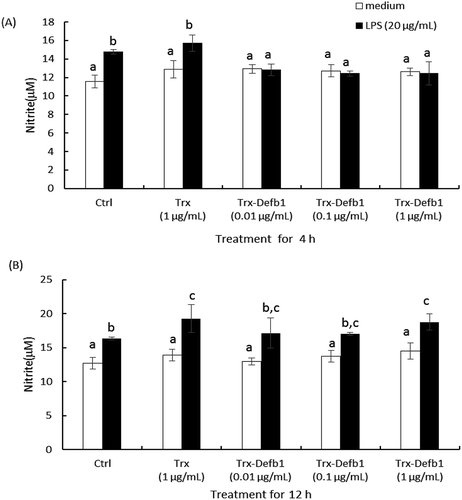
To evaluate the immuno-regulatory role of gcDefb1, the effect of Trx-Defb1 on the secretion of gcTnf-α was examined in HKM upon LPS stimulation. As shown in , Trx-Defb1 (1 μg/mL) significantly down-regulated LPS-induced gcTnf-α secretion compared with control groups (LPS alone and LPS combined with Trx). This effect was detected as early as 4 h after treatment ()) and disappeared at 12 h ()). Additionally, Trx-rgcDefb1 was not effective in mediating basal gcTnf-α release in the same cell model (Data not shown). Subsequently, the effect of Trx-Defb1 on the production of LPS-induced NO was assayed. As shown in , compared with control groups (LPS alone or LPS combined with Trx), Trx-Defb1 down-regulated LPS-stimulated nitric oxide production in a dose-dependent manner (0.01–1 μg/mL, )). The response disappeared 12 h post treatment ()). Meanwhile, Trx at the same concentration did not affect the effect of LPS on nitric oxide production, and Trx-Defb1 had no effect on basal levels of nitric oxide in HKM.
Figure 5. Effect of Trx-Defb1 on LPS-induced gcTnf-α secretion in grass carp HKM.
Grass carp HKM were exposed to Trx-Defb1 (0.1 and 1 μg/mL) in the presence of LPS (20 μg/mL) for 4 h (A) and 12 h (B). The gcTnf-α in the supernatant was measured by ELISA. Trx was set as the control. Data presented are expressed as Mean ± SEM (N = 4) which are the representative results from three individual experiments. The alphabet denotes the significant differences at p < 0.05.
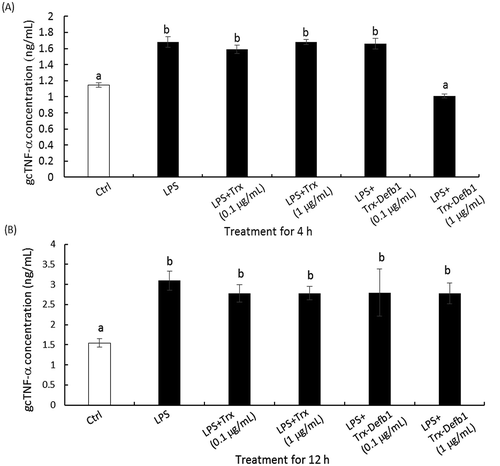
Figure 6. Effect of Trx-Defb1 on LPS-induced nitric oxide production in grass carp HKM.
The cells were exposed to increasing concentrations of Trx-Defb1 (0.01 to 1 μg/mL) in the presence of medium or LPS (20 μg/mL) for 4 h (A) and 12 h (B). NO in the supernatant was measured by Griess reaction. Trx was set as the control. Data presented are expressed as Mean ± SEM (N = 4) which are the representative results from three individual experiments. The alphabet denotes the significant differences at p < 0.05.
In this study, the inhibitory effects of gcDefb1 on inflammatory response, together with that gcdefb1 displayed a ubiquitous expression pattern in the selected tissues, implied the potential of gcDefb1 in maintaining noninflammatory homeostasis. Given that teleosts frequently contact numerous microbes in a complex aquatic environment, our findings reinforced the role of β-defensin 1 in fish inflammatory control depending on inhibition of both pathogens and inflammation.
Author contribution
Conceived and designed the experiments: Taiqiang Zhao, Kun Yang and Fangfang Ren. Performed the experiments: Boren Hou and Kun Yang. Analyzed the data: Taiqiang Zhao, Kun Yang and Fangfang Ren. Wrote the paper: Taiqiang Zhao, Kun Yang and Hong Zhou.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Zhao JG, Zhou L, Jin JY, et al. Antimicrobial activity-specific to Gram-negative bacteria and immune modulation-mediated NF-κB and Sp1 of a medaka β-defensin. Dev Comp Immunol. 2009;33(4):624–637.
- Zhu J, Wang H, Wang J, et al. Identification and characterization of a β-defensin gene involved in the immune defense response of channel catfish, Ictalurus punctatus. Mol Immunol. 2017;85:256–264.
- Dong JJ, Wu F, Ye X, et al. β-Defensin in Nile tilapia (Oreochromis niloticus): sequence, tissue expression, and anti-bacterial activity of synthetic peptides. Gene. 2015;566(1):23–31.
- Ruangsri J, Kitani Y, Kiron V, et al. A novel beta-defensin antimicrobial peptide in Atlantic cod with stimulatory effect on phagocytic activity. PLoS One. 2013;8(4):e62302.
- Semple F, Dorin JR. beta-Defensins: multifunctional modulators of infection, inflammation and more? J Innate Immun. 2012;4(4):337–348.
- Funderburg N, Lederman MM, Feng Z, et al. Human β-defensin-3 activates professional antigen-presenting cells via Toll-like receptors 1 and 2. Proc Natl Acad Sci USA. 2007;104(47):18631–18635.
- Tani K, Murphy WJ, Chertov O, et al. Defensins act as potent adjuvants that promote cellular and humoral immune responses in mice to a lymphoma idiotype and carrier antigens. Int Immunol. 2000;12(5):691–700.
- Hancock REW, Haney EF, Gill EE. The immunology of host defence peptides: beyond antimicrobial activity. Nat Rev Immunol. 2016;16(5):321–334.
- Wang Y, Lu Y, Zhang Y, et al. The draft genome of the grass carp (Ctenopharyngodon idellus) provides insights into its evolution and vegetarian adaptation. Nat Genet. 2015;47(6):625–631.
- Song X, Zhao J, Bo Y, et al. Aeromonas hydrophila induces intestinal inflammation in grass carp (Ctenopharyngodon idella): an experimental model. Aquaculture. 2014;434(SupplementC):171–178.
- Wang G, Li X, Wang Z. APD3: the antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 2016;44(D1):D1087–D1093.
- Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–1874.
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406–425.
- Harder J, Bartels J, Christophers E, et al. Isolation and characterization of human beta -defensin-3, a novel human inducible peptide antibiotic. J Biol Chem. 2001;276(8):5707–5713.
- Nam BH, Moon JY, Kim YO, et al. Multiple β-defensin isoforms identified in early developmental stages of the teleost Paralichthys olivaceus. Fish Shellfish Immunol. 2010;28(2):267–274.
- Yang K, Zhang S, Chen D, et al. IFN-γ-activated lymphocytes boost nitric oxide production in grass carp monocytes/macrophages. Fish Shellfish Immunol. 2013;35(5):1635–1641.
- Zhang A, Chen D, Wei H, et al. Functional characterization of TNF-alpha in grass carp head kidney leukocytes: induction and involvement in the regulation of NF-kappaB signaling. Fish Shellfish Immunol. 2012;33(5):1123–1132.
- Green LC, Wagner DA, Glogowski J, et al. Analysis of nitrate, nitrite, and [15N]nitrate in biological fluids. Anal Biochem. 1982;126(1):131–138.
- Marel M, Adamek M, Gonzalez SF, et al. Molecular cloning and expression of two beta-defensin and two mucin genes in common carp (Cyprinus carpio L.) and their up-regulation after beta-glucan feeding. Fish Shellfish Immunol. 2012;32(3):494–501.
- Casadei E, Wang T, Zou J, et al. Characterization of three novel β-defensin antimicrobial peptides in rainbow trout (Oncorhynchus mykiss). Mol Immunol. 2009;46(16):3358–3366.
- Lai Y, Gallo RL. AMPed up immunity: how antimicrobial peptides have multiple roles in immune defense. Trends Immunol. 2009;30(3):131–141.
- Zou J, Mercier C, Koussounadis A, et al. Discovery of multiple beta-defensin like homologues in teleost fish. Mol Immunol. 2007;44(4):638–647.
- Soscia SJ, Kirby JE, Washicosky KJ, et al. The Alzheimer’s disease-associated amyloid beta-protein is an antimicrobial peptide. PLoS One. 2010;5(3):e9505.
- Wang T, Secombes CJ. The cytokine networks of adaptive immunity in fish. Fish Shellfish Immunol. 2013;35(6):1703–1718.
- Ohshima H, Bartsch H. Chronic infections and inflammatory processes as cancer risk factors: possible role of nitric oxide in carcinogenesis. Mutat Res. 1994;305(2):253–264.
