ABSTRACT
We evaluated the import of Arabidopsis catalase to peroxisomes under homogenous transient expression. The amino acids at −11 to −4 from the C-terminus are necessary for catalase import. The results are in agreement with the previous work under stable expression. We first demonstrate that heme-binding sites are important for peroxisomal import, suggesting the importance of catalase folding.
Abbreviations: AtCat: Arabidopsis catalase; PTS: peroxisomal targeting signal; PEX: Peroxin
Catalase is a tetrameric heme-containing enzyme scavenging hydrogen peroxide accumulated in peroxisomes. Peroxisomal proteins, including catalase, are imported into peroxisomes, based on either of two types of signal sequence called the peroxisomal targeting signal (PTS), PTS1 and PTS2 [Citation1]. A typical PTS1 is based on a consensus tripeptide, SKL, at C-terminus. The existence of its variants with a consensus tripeptide, S/A/C-K/R/H-L/M, has also been reported [Citation2,Citation3]. In contrast to the PTS1, the PTS2 is located within about 30 amino acids from N-terminus [Citation1,Citation4]. During the import to peroxisomes, the PTS1 and PTS2 are recognized by peroxisomal biogenesis factors, Peroxin 5 (PEX5) and PEX7 respectively. Plant catalase appears to have neither a typical PTS1 nor PTS2. In cotton seed, the three C-terminal amino acids (PSI) of catalase are necessary for the import to peroxisomes[Citation5]. Similarly, a tripeptide (P-S/T-I/M) at C-terminus is conserved in various plant catalases. Therefore, the three C-terminal amino acids may function as PTS1. On the other hand, we found previously that three amino acids (PSI) from the C-terminus of pumpkin catalase, Cat1, were not required for catalase import [Citation6], although a yeast two-hybrid showed that pumpkin Cat1 interacted with PEX5. Instead of PSI, pumpkin Cat1 has QKL at the positions 13 to 11 from the C-terminus, which shows similar molecular characteristics to the SKL motif. Therefore, the tripeptide at the positions 13 to 11 might function as an internal PTS1. However, catalase import remains unclear.
We have evaluated the import of pumpkin Cat1 to peroxisomes in tobacco and Arabidopsis by stable expression [Citation6]. However, it takes time to obtain results under stable expression. In this study, we transiently expressed catalase as a green fluorescent protein (GFP) fusion protein in Arabidopsis protoplasts using the method by Fujikawa et al [Citation7]. To avoid the possibility that the variation in import machinery is due to heterogeneous catalase, we used Arabidopsis catalase, instead of pumpkin Cat1. Upon comparison by the alignment of three Arabidopsis catalase: AtCat1, AtCat2 and AtCat3 with pumpkin Cat1, AtCat2 has the highest similarity (91% identity) with pumpkin Cat1. Two tripeptides, PSI and QKL, are consistently found at the C-terminal end and the positions 13 to 11 from the C-terminus of AtCat2, respectively. We fused the DNA fragment coding AtCat2 (GenBank Accession No. NP195234) at the 5′- and 3′- ends of the GFP coding sequence. We expressed under the CaMV35S promoter (35S), with the peroxisomal marker, RFP-PTS1 [Citation8], which is the fusion protein of red fluorescent protein (RFP) with a consensus PTS1 (SKL). The empty vector, p35S::GFP, was used as a control. Subcellular localizations of GFP and RFP-PTS1 were analyzed with a confocal laser scanning microscope (LMS 700; Carl Zeiss). The green fluorescent signals from the control were detected in the nucleus and cytosol ()), while the red fluorescent signals from RFP-PTS1, which are present as red spots, were detected only in the peroxisomes. For GFP-AtCat2, its GFP signals were observed as dot patterns and overlapped with the co-expressed RFP signals, indicating the peroxisomal localization of GFP-AtCat2. When GFP was fused at the C-terminus (AtCat2-GFP), similar results were obtained. The detection of GFP signals from AtCat2-GFP in the peroxisomes suggests that GFP fusion at the C-terminus did not mask PTS. In Amaranthus cruentus, a catalase-phenol oxidase did not localize to the peroxisome as revealed by fusing the GFP at its C-terminus [Citation9]. To our knowledge, there are no reports on the subcellular localization of plant catalase fused with GFP at C-terminus. In addition, the GFP signals from GFP-PTS1 and GFP-AtCat2 were observed in the cytosol when RFP-AtPEX5 was co-expressed, which may be caused by disorders of PEX5 import machinery with the excessive expression or the dominant-negative form of PEX5. Similarly, the pumpkin Cat1 could not be imported into peroxisomes in Arabidopsis knockdown mutant of the AtPEX5 gene [Citation10]. Therefore, the results suggest that the AtCat2 has an internal PTS1 recognized by AtPEX5.
Figure 1. The contribution of the C-terminus of Arabidopsis Cat2 to its subcellular localization.
Arabidopsis protoplasts were co-transfected with plant expression vectors under control of the constitutive CaMV35S promoter with the sequence coding Arabidopsis AtCat2 fused with GFP and peroxisomal marker; RFP-PTS1 or RFP-AtPEX5. The signals of GFP (left) and RFP (middle) were observed, and they were merged (right). (a) Upper three-row panels; GFP only (GFP) or GFP fused at the N-terminus (GFP-AtCat2) and C-terminus (AtCat2-GFP) of AtCat2 were transiently expressed with RFP-PTS1. Lower two-row panels; GFP-PTS1 or GFP-AtCat2 was transiently expressed with RFP-AtPEX5. (b) Upper part shows the representation of the C-terminal amino acids of AtCat2 and its mutants. Lower part shows the signals of GFP and RFP in the transfected protoplasts. The amino acids (QKLA) of GFP-AtCat2 at the positions 480 to 483 from the N-terminus of AtCat2 were substituted to Gly (Q480G, K481G, L482G and A483G, respectively). (c) Upper part shows the representation of the C-terminal amino acids of AtCat2 and its truncated mutants. Lower part shows the signals of GFP and RFP in the transfected protoplasts. The C-terminal 3, 9, 10 and 11 amino acids of AtCat2 were deleted from GFP-AtCat2 (ΔC3, ΔC9, ΔC10 and ΔC11, respectively). Scale bars = 5 µm.
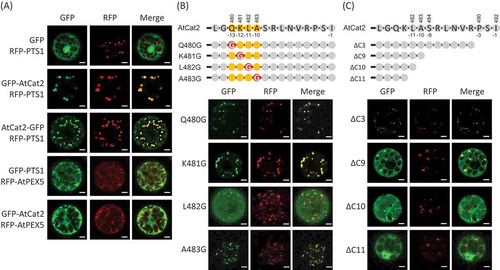
The C-terminal position 11 of pumpkin Cat1 was important for the import to peroxisomes [Citation6]. In this study, we examined the function of QKLA at the positions 13 to 10 by substitution of the tetrapeptide to Gly ()). The GFP signals were observed in peroxisomes when GFP was fused to three mutated AtCat2 (Q480G, K481G and A483G). However, GFP signal was observed in the cytosol when the Leu was substituted (L482G), showing that the Leu is required for the import of AtCat2 to peroxisomes. To further validate the contribution of the C-terminus to this localization, we expressed GFP fused to AtCat2 lacking the C-terminus (ΔC3, ΔC9, ΔC10, and ΔC11; )). When the C-terminal tripeptide was deleted, the GFP signal of the GFP-AtCat2ΔC3 was observed at the peroxisomes. When 9 and 10 amino acids were deleted, these GFP signals were confined to the peroxisomes and the cytosol. Moreover, when the GFP fused to AtCat2 lacking the 11 C-terminal amino acids was expressed, its GFP signal was confined only to the cytosol, and no more GFP signal was detected in the peroxisomes. These results suggest that the amino acids at the positions 11 to 4, especially position 11, are involved in the import of AtCat2 to peroxisomes. We previously reported that pumpkin Cat1 lacking C-terminal tripeptides was not imported into peroxisome by transient expression in tobacco and Arabidopsis [Citation6], in contrast with its stable expression and the current result. The import of pumpkin Cat1 to peroxisomes under transient expression in heterogeneous species may have low efficiency, compared with that under homogeneous expression.
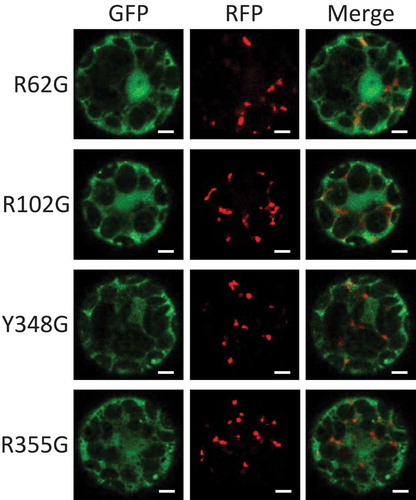
Catalase is a heme-containing enzyme. In heme-containing proteins, the heme binding should support protein folding [Citation11,Citation12]. However, there are not any direct evidences whether heme binding sites are involved in the import of plant catalase to peroxisomes. To evaluate the role of amino acids related to heme binding in catalase import, mutations were designed to substitute the amino acids of heme-binding sites. Three Arg at the positions 62, 102, and 355 and a Tyr at the position 348 from the N-terminus appear to be involved in heme binding, compared with human catalase [Citation13]. We examined the effect of substitution of these amino acids with Gly by observation of the subcellular localization of the GFP fused to these mutated AtCat2 (). All the GFP signals from these mutated AtCat2 (R62G, R102G, Y348G, and R355G) were observed in the cytosol and around the nucleus, indicating that the amino acids of heme-binding sites are essential for catalase import. Proteins with the SKL motif conjugated with gold particles were imported into peroxisomes [Citation14]. The report indicates that peroxisomal proteins are imported as the large complex consisting of folded proteins. Therefore, even though catalase is a tetramer, it can be imported as the large complex. In this study, we did not evaluate the heme binding and folding of AtCat2. However, these mutations must affect protein folding. The misfolding of AtCat2 may cause difficulty in displaying PTS1 at the C-terminus to PEX5.
Figure 2. The role of the amino acids related to heme binding of Arabidopsis Cat2 in its subcellular localization.
Arabidopsis AtCat2 with GFP fused at the N-terminus and RFP-PTS1 were transiently expressed under control of constitutive CaMV35S promoter in Arabidopsis protoplasts. The transfected protoplasts were observed for signals of GFP (left) and RFP (middle), and they were merged (right). The four amino acids (positions 62, 102, 348 and 355 from the N-terminus of AtCat2) related to heme binding were substituted with Gly (R62G, R102G, Y348G and R355G, respectively). Scale bars = 5 µm.
In this study, we evaluated the import of Arabidopsis catalase to peroxisomes under homogeneous transient expression. The previous work showed the different results from transient expression and stable expression systems in heterogenous protein species [Citation6]. Heterogenous expression may affect the efficiency of catalase import. Considering that the current results are in good agreement with the results under stable expression [Citation6], the homogenous transient expression is effective for analyzing the import of plant catalase to peroxisomes. Also, transient expression is a time-saving technique for evaluating the import of mutated proteins to peroxisomes. The current homogenous transient expression showed, in Arabidopsis AtCat2, at least the amino acids at the positions 11 to 4 from the C-terminus are considered as contributing catalase import. These eight amino acid residues contain an SKL motif-like tripeptide (SRL). The Leu at the position 11 from the C-terminus is necessary for catalase import. Besides, we demonstrated for the first time that the amino acids related to heme binding (three Arg residues at the positions 62, 102, and 355 and Tyr at the position 348) contribute to the import of Arabidopsis catalase to peroxisomes, as well as the C-terminus residues. Heme binding is attributed to catalase folding. The folding of catalase may be important for the import to peroxisomes. Therefore, the elucidation of the relationship between folding and targeting for catalase import will be required to shed light on the import of plant catalase to peroxisomes.
Author contribution
Fujikawa and Suekawa designed research and wrote the paper. Endo and Fukami performed the experiments. Mano and Nishimura gave the advice of research and paper. Esaka designed research and gave the advice of paper.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Baker A, Lanyon-Hogg T, Warriner SL. Peroxisome protein import: a complex journey. Biochem Soc Trans. 2016;44(3):783–789.
- Brocard C, Hartig A. Peroxisome targeting signal 1: is it really a simple tripeptide? Biochim Biophys Acta. 2006;1763(12):1565–1573.
- Williams C, Bener Aksam E, Gunkel K, et al The relevance of the non-canonical PTS1 of peroxisomal catalase. Biochim Biophys Acta. 2012;1823(7):1133–1141.
- Petriv OI, Tang L, Titorenko VI, et al A new definition for the consensus sequence of the peroxisome targeting signal type 2. J Mol Biol. 2004;341(1):119–134.
- Mullen RT, Lee MS, Trelease RN. Identification of the peroxisomal targeting signal for cottonseed catalase. Plant J. 1997;12(2):313–322.
- Kamigaki A, Mano S, Terauchi K, et al Identification of peroxisomal targeting signal of pumpkin catalase and the binding analysis with PTS1 receptor. Plant J. 2003;33(1):161–175.
- Fujikawa Y, Kato N. Split luciferase complementation assay to study protein-protein interactions in Arabidopsis protoplasts. Plant J. 2007;52(1):185–195.
- Mano S, Nakamori C, Nito K, et al The Arabidopsis pex12 and pex13 mutants are defective in both PTS1- and PTS2-dependent protein transport to peroxisomes. Plant J. 2006;47(4):604–618.
- Chen N, Teng XL, Xiao XG. Subcellular localization of a plant catalase-phenol oxidase, AcCATPO, from Amaranthus and identification of a non-canonical peroxisome targeting signal. Front Plant Sci. 2017;8:1345.
- Oshima Y, Kamigaki A, Nakamori C, et al Plant catalase is imported into peroxisomes by Pex5p but is distinct from typical PTS1 import. Plant Cell Physiol. 2008;49(4):671–677.
- Smith LJ, Kahraman A, Thornton JM. Heme proteins–diversity in structural characteristics, function, and folding. Proteins. 2010;78(10):2349–2368.
- Janosik M, Oliveriusova J, Janosikova B, et al Impaired heme binding and aggregation of mutant cystathionine beta-synthase subunits in homocystinuria. Am J Hum Genet. 2001;68(6):1506–1513.
- Diaz A, Loewen PC, Fita I, et al Thirty years of heme catalases structural biology. Arch Biochem Biophys. 2012;525(2):102–110.
- Walton PA, Hill PE, Subramani S. Import of stably folded proteins into peroxisomes. Mol Biol Cell. 1995;6(6):675–683.
