ABSTRACT
Endoplasmic reticulum (ER)-located protein Ire1 triggers the unfolded protein response against ER-stressing stimuli, which are categorized as ER accumulation of unfolded proteins or membrane lipid-related aberrancy. Here we demonstrate that by using yeast Ire1 mutants, we can distinguish the category to which a stress-inducing stimulus belongs. For instance, ethanol was found to activate Ire1 through both types of cellular damage.
The endoplasmic reticulum (ER) is a membrane-bound cellular compartment where secretory and transmembrane proteins are folded. Moreover, membrane-lipid components are mainly biosynthesized on the ER. An early study on mammalian cells found that stress-inducing stimuli that impair protein folding in the ER commonly induce genes encoding molecular chaperones and protein folding enzymes located in the ER [Citation1]. While this cellular response, now termed the ER stress response or the unfolded protein response (UPR), is observable throughout eukaryotic species, its mechanism was initially uncovered through yeast studies. In Saccharomyces cerevisiae cells, ER-located transmembrane protein Ire1 acts as an endoribonuclease and contributes to the cytoplasmic splicing of the HAC1 mRNA, which is then translated into a transcription factor protein responsible for the UPR [Citation2]. UPR target genes include those encoding enzymes for membrane lipid metabolism as well as those encoding factors to cope with unfolded proteins that accumulate in the ER [Citation3,Citation4].
Cellular conditions that induce the UPR are collectively called ER stress. Accumulation of unfolded proteins in the ER is a major type of ER stress and is thought to be directly detected by Ire1, since, according to previous studies, the luminal domains of S. cerevisiae Ire1 and IRE1α (the major Ire1 paralogue in mammalian cells) directly capture unfolded proteins, leading to their oligomerization and activation [Citation2,Citation5–Citation9]. On the other hand, disturbance of membrane lipid homeostasis is likely to be another type of ER stress that activates Ire1 independently of the accumulation of unfolded proteins in the ER [Citation7,Citation10]. Ref [Citation11]. reported that the transmembrane domain of Ire1 has a unique property that causes oligomerization and activation of Ire1 in a manner dependent on membrane lipid composition. In the case of S. cerevisiae Ire1, this feature is likely to be impaired by transmembrane domain point mutations F531R and V535R [Citation11].
These insights can expand to a question: which mediates induction of the UPR by an external stimulus, ER accumulation of unfolded proteins or disturbance of membrane lipid homeostasis? As exemplified by the case of exposure of S. cerevisiae cells to ethanol, conflicting observations have often been presented. We previously reported that ethanol denatures ER client proteins, which then trigger the UPR [Citation12]. On the other hand, according to Ref [Citation13]., ethanol increases membrane fluidity, leading to induction of the UPR.
Ire1 mutants that selectively compromise the UPR by one of these two categories of ER stress will provide important clues to understanding the mechanism that leads to induction of the UPR. We previously reported that the ability of S. cerevisiae Ire1 to capture unfolded proteins is impaired by deletion of its amino-acid residues 253–272 (∆III mutation), which are corresponding to the full-length Subregion III, a loosely folded segment located inside the unfolded protein-capturing module of Ire1 [Citation5–Citation7,Citation14]. According to our previous report [Citation7], the ∆III mutation lowers the activity of Ire1 induced by ER accumulation of unfolded proteins but not by membrane lipid-related aberrancy. On the other hand, while the F531R and the V535R point mutations are expected to specifically compromise Ire1 activation through membrane lipid-related aberrancy, to our knowledge, such an observation has yet to be reported.
In this study, we examined whether, as expected, mutations in S. cerevisiae Ire1 that specifically impair its responsiveness to either unfolded proteins or membrane lipid-related aberrancy actually change its activation profile in response to different kinds of ER stressing stimuli. S. cerevisiae ire1∆ strains W303-ire1∆ (W303 background) or KMY1516 (SEY6210 background) transformed with the YCp-type plasmid pRS313-IRE1, which carries the wild-type IRE1 gene, were used as the wild-type IRE1 strains through this study [Citation14,Citation15]. ∆III and V535R mutations were introduced into pRS313-IRE1 using in vivo homologous recombination [Citation6,Citation14]. Cells were exponentially grown by shaking in standard dextrose (SD) medium at 30 °C, then stressed as indicated in the figure legends. Total RNA samples were then used for reverse transcription-PCR analysis to amplify the HAC1 transcripts [Citation7]. The resulting data was used to estimate the ratio of spliced HAC1 mRNA to total (spliced and unspliced) HAC1 mRNA [Citation7], which is termed the HAC1 mRNA-splicing efficiency and serves as the index of Ire1 activity in cells.
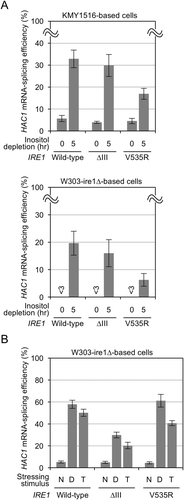
When wild-type IRE1 cells and the cells carrying ∆III Ire1 were stressed by inositol depletion, HAC1 mRNA splicing was induced almost equally in both cells, as reported previously [Citation7,Citation11] and shown in . Although the W303 background strains (W303-ire1∆ derivatives) are less sensitive to inositol depletion than the SEY6210 background strains (KMY1516 derivatives), these two types of strains exhibited a similar pattern of results (). The experiment shown in also reproduced our previous observation that the ∆III mutation compromises HAC1-mRNA splicing and Ire1 activation induced by cellular treatment with tunicamycin or dithiothreitol (DTT) [Citation6,Citation7]. This observation is consistent with a commonly acceptable expectation that the N-glycosylation inhibitor tunicamycin and the disulfide reducing agent DTT primarily damage protein folding in the ER. On the other hand, depletion of inositol, which is an important membrane lipid constituent, is deduced to induce the UPR through membrane lipid-related aberrancy. As reported in Ref [Citation11]. and shown in , the V535R mutation compromised HAC1 mRNA splicing following inositol depletion. Here we newly show that, at least under the conditions used in this study, cellular treatment with tunicamycin or DTT almost equally induced HAC1 mRNA splicing in cells with wild-type and V535R Ire1 ().
Figure 1. Splicing of the HAC1 mRNA by wild-type, V535R, and ∆III Ire1 in S. cerevisiae following treatment with conventional ER-stressing stimuli. (a) KMY1516 (ire1∆) cells or W303-ire1∆ (ire1∆) cells transformed with pRS313-IRE1 or its indicated mutants were stressed by culturing in inositol-depleted SD medium. (b) W303-ire1∆ (ire1∆) cells transformed with pRS313-IRE1 or its indicated mutants were cultured with the indicated chamicals. N: non-stress, D: 10 mM DTT for 30 min, and T: 2 µg/ml tunicamycin for 1 hr.
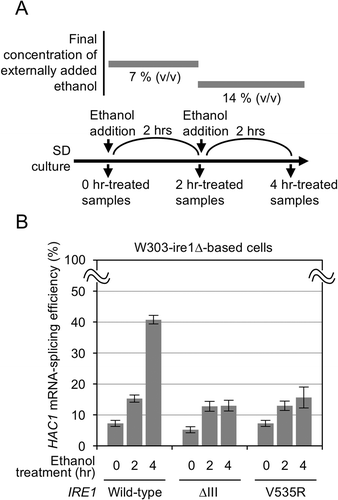
These findings are highly supportive of the aforementioned theory presented by us and others that ER accumulation of unfolded proteins and membrane lipid-related aberrancy are detected by Ire1 by distinct mechanisms [Citation7,Citation8,Citation10,Citation11]. Moreover, we anticipated that we can clearly distinguish these two types of ER stress by using the ∆III and the V535R mutants of S. cerevisiae Ire1. We thus addressed the activation mechanism of Ire1 following ethanol treatment of S. cerevisiae cells, which, as aforementioned, has yet to be concluded [Citation12,Citation13]. Here cells were stressed by stepwise addition of ethanol into medium as done previously (, Ref [Citation12]). shows that both the ∆III and the V535R mutations compromised HAC1 mRNA splicing induced by ethanol stress. Thus, we propose that ethanol stress activates Ire1 through both types of cellular damage.
Figure 2. Splicing of the HAC1 mRNA by wild-type, V535R, and ∆III Ire1 in S. cerevisiae stressed by ethanol. (a) Culturing procedure for ethanol treatment of S. cerevisiae cells. (b) W303-ire1∆ (ire1∆) cells transformed with pRS313-IRE1 or its indicated mutants were stressed by ethanol as shown panel A.
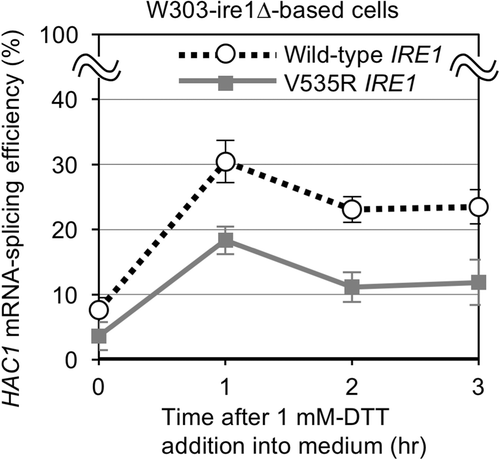
Ref [Citation11]. showed a reduction in activity of S. cerevisiae Ire1 following DTT stress by the V535R mutation, which is inconsistent with our result shown in . We hypothesized that this discrepancy is due to differences in experimental conditions, such as the strength of DTT imposition. To validate this hypothesis, we examined HAC1 mRNA splicing in response to cellular treatment with a lower concentration of DTT. As shown in , treatment with 1 mM DTT induced HAC1 mRNA splicing weaker in cells with V535R Ire1 than wild-type IRE1 cells. We thus propose the following scenario. Strong DTT stress produces a large amount of unfolded proteins, which is abundant enough to activate V535R Ire1 as well as wild-type Ire1 (). In other words, although it is also induced, membrane lipid-related aberrancy is not the determinant of activity of Ire1 under strong DTT conditions. Conversely, under weaker DTT stress conditions, unfolded proteins accumulate in the ER less abundantly, so that membrane lipid-related aberrancy greater contributes to Ire1 activation.
Figure 3. Splicing of the HAC1 mRNA by wild-type and V535R Ire1 in S. cerevisiae cultured with dilute DTT. W303-ire1∆ (ire1∆) cells transformed with pRS313-IRE1 or its V535R mutant were cultured with 1 mM DTT for the indicated durations.
In conclusion, here we showed a distinction between the two types of ER stress using S. cerevisiae Ire1 mutants. As it is now likely that mammalian IRE1α also detects these two types of ER stress in a similar manner to S. cerevisiae Ire1 [Citation9,Citation10], our methodology could be also applied to mammalian studies.
Author contribution
Y.K. managed the project and wrote the manuscript. D.M.T. and Y.K. carried out the experiments. H.T. provided valuable and indispensable advice on the study.
Acknowledgments
We deeply thank to Prof. Kenji Kohno (Nara Inst. Sci. Tech.) for a wide variety of valuable supports, which have been indispensable for this study.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Kozutsumi Y, Segal M, Normington K, et al The presence of malfolded proteins in the endoplasmic reticulum signals the induction of glucose-regulated proteins. Nature. 1988;332:462–464.
- Kimata Y, Kohno K. Endoplasmic reticulum stress-sensing mechanisms in yeast and mammalian cells. Curr Opin Cell Biol. 2011;23:135–142.
- Travers KJ, Patil CK, Wodicka L, et al Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000;101:249–258.
- Kimata Y, Ishiwata-Kimata Y, Yamada S, et al Yeast unfolded protein response pathway regulates expression of genes for anti-oxidative stress and for cell surface proteins. Genes Cells. 2006;11:59–69.
- Credle JJ, Finer-Moore JS, Papa FR, et al On the mechanism of sensing unfolded protein in the endoplasmic reticulum. Proc Natl Acad Sci USA. 2005;102:18773–18784.
- Kimata Y, Ishiwata-Kimata Y, Ito T, et al Two regulatory steps of ER-stress sensor Ire1 involving its cluster formation and interaction with unfolded proteins. J Cell Biol. 2007;179:75–86.
- Promlek T, Ishiwata-Kimata Y, Shido M, et al Membrane aberrancy and unfolded proteins activate the endoplasmic reticulum stress sensor Ire1 in different ways. Mol Biol Cell. 2011;22:3520–3532.
- Gardner BM, Walter P. Unfolded proteins are Ire1-activating ligands that directly induce the unfolded protein response. Science. 2011;333:1891–1894.
- Karagoz GE, Acosta-Alvear D, Nguyen HT, et al. An unfolded protein-induced conformational switch activates mammalian IRE1. eLife. 2017;6.
- Volmer R, van der Ploeg K, Ron D. Membrane lipid saturation activates endoplasmic reticulum unfolded protein response transducers through their transmembrane domains. Proc Natl Acad Sci USA. 2013;110:4628–4633.
- Halbleib K, Pesek K, Covino R, et al Activation of the unfolded protein response by lipid bilayer stress. Mol Cell. 2017;67:673–684.
- Miyagawa K, Ishiwata-Kimata Y, Kohno K, et al Ethanol stress impairs protein folding in the endoplasmic reticulum and activates Ire1 in Saccharomyces cerevisiae. Biosci Biotechnol Biochem. 2014;78:1389–1391.
- Navarro-Tapia E, Querol A, Pérez-Torrado R. Membrane fluidification by ethanol stress activates unfolded protein response in yeasts. Microb Biotechnol. 2018;11:465–475.
- Kimata Y, Oikawa D, Shimizu Y, et al A role for BiP as an adjustor for the endoplasmic reticulum stress-sensing protein Ire1. J Cell Biol. 2004;167:445–456.
- Le QG, Ishiwata-Kimata Y, Kohno K, et al Cadmium impairs protein folding in the endoplasmic reticulum and induces the unfolded protein response. FEMS Yeast Res. 2016;16:fow049.
