ABSTRACT
MiRNA-5195-3p (miR-5195-3p), a recently discovered and poorly studied miRNA, has been reported to suppress bladder cancer cell behavior. However, its regulatory role in non-small cell lung cancer (NSCLC) remains unclear. Here, the expression of miR-5195-3p was found to be reduced in NSCLC tissues and cells. The in vitro experiments showed that miR-5195-3p upregulation repressed cell proliferation, migration and invasion by CCK-8 and transwell assays. In addition, MYO6 was predicted and confirmed as a potential target of miR-5195-3p by Bioinformatics analysis, Luciferase reporter assay and western blot analysis. There was significantly negative correlation between miR-5195-3p and MYO6 in NSCLC tissues. Furthermore, MYO6 knockdown exhibited similar effects to those of miR-5195-3p overexpression in NSCLC cells, and restored MYO6 expression reversed the inhibitory effects of miR-5195-3p. Therefore, these results demonstrate that miR-5195-3p functions as a tumor suppressor by directly modulating MYO6 expression in NSCLC cells, and may be an innovative candidate target for NSCLC therapy.
Graphical Abstract
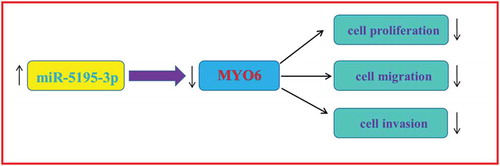
MicroRNA-5195-3p plays a suppressive role in cell proliferation, migration and invasion by targeting MYO6 in human non-small cell lung cancer.
Non-small cell lung cancer (NSCLC) is a highly heterogeneous disease, represents ~ 85% of all newly diagnosed lung cancers [Citation1]. Tobacco smoking is a major risk factor for developing NSCLC. Additionally, genetic, epigenetic, and environmental factors (such as asbetos and radon) also have a role [Citation2,Citation3]. So far, the majority of patients are diagnosed at late stage largely attributable to delayed onset of clinical features and inadequate screening technologies [Citation4]. Although advanced patients benefit from the standard management strategy, the combination of platium-based chemotherapy and second-line cytotoxic chemotherapy, patients still have very poor prognosis with a median survival time of approximately 1 year. Greatly deepen understanding of progression of NSCLC especially molecular biology is fundamental to hasten the development of treatment.
Accumulating studies revealed that tumor initiation, growth and metastasis can involve microRNAs (miRNAs), a class of short (19 to 24 nt) non-coding RNAs which dysregulation or mutation are ubiquitous in human malignancies [Citation5]. The first evidence that miRNAs-cancer connection derived from a previous report characterizing the loss of 13q14 (including two miRNA members miR-15a and miR-16–1) in more than 50% of all chronic lymphocytic leukemia cases [Citation6]. Functionally, a variety of miRNAs probably facilitate tumorigenesis primarily through modulating the tumor suppressors, as is the sample for miR-21, which post-transcriptionally downregulates Pdcd4 in colorectal cancer [Citation7], and miR-155, which targeting VHL tumor suppressor in breast cancer [Citation8]. Besides, many investigations highlight an anti-oncogene role for miRNAs in carcinogenesis, as in the case for miR-185, which transcriptionally depress Six1 in breast cancer, ovarian and pediatric renal tumors [Citation9], and miR-194 prevents aggressive behavior of endometrial cancer cells though downregulates a potent oncogene BMI-1 [Citation10]. MiR-5195-3p is low-expressed in bladder cancer cell lines and regulates their proliferation and invasion via repression of oncogene KLF5 [Citation11]. Zhang et al. [Citation12] revealed that miR-5195-3p involves downregulation of Beclin1 expression mediated by overexpression of p72, and contribute to glioma cells migration and invasion. However, the direct evaluation of the links between miR-5195-3p and human malignancies is still rare, let alone in the field of NSCLC.
As an unconventional family of myosin molecules, Myosin VI (MYO6) is widely expressed in various organisms and tissue types [Citation13]. The protein acts as an anchor and a transporter, hydrolyse ATP to moves toward the minus ends of actin filaments, unlike other known myosins [Citation14]. It was subsequently found to play crucial roles in maintenance of stereocilia structure, vesicle trafficking, endocytosis, epithelial morphogenesis, and autophagosome maturation [Citation15]. Moreover, disruption of these processes has been linked to deafness, cancers, neurodegeneration and hypertropic cardiomyopathy in the human population [Citation16]. It is well established that mutation of MYO6 involved in recessive and dominant forms of human nonsyndromic deafness [Citation17], defects in the human homologue of the Snell’s waltzer mice gene could also resulting in hearing loss in mice [Citation18]. In clinical prostate specimens, MYO6 was identified as one of the top enriched gene, and silencing of the gene was accompanied by impairment of migration and colony formation [Citation19].
Until now, there has been no study revealing whether MYO6 is being controlled by miRNAs in NSCLC. This study was undertaken (1) to determine the expression profiles of miR-5195-3p and MYO6 in NSCLC tissues and/or cell lines, (2) to evaluate the regulation of MYO6 by miR-5195-3p and the corresponding mechanisms that mediate the regulation, (3) to reveal the role of the two genes in NSCLC cells proliferation, invasion and migration. This study may provide a novel theoretical and experimental basis for exploitation of therapeutic strategy.
Materials and methods
Tissue samples and cell lines
A total of 30 paired human NSCLC tissues and the matched adjacent non-tumorous lung tissues were collected from patients who underwent surgical resection at The First Hospital of Tianshui (Gansu, China). Before received surgery, all patients did not undergo chemotherapy or radiotherapy. The patients’ medical records, including age, sex, tumor size, histological grade, Tumor Node Metastasis (TNM) stage and lymph node metastasis were summarized in . The histological grade was classified according to the Four-tier grading scheme as previously described [Citation20]. TNM stage was classified based on the 8th edition of the Union for International Cancer Control (UICC)-TNM classification [Citation21]. The resected lung tissues were immediately snap-frozen in liquid nitrogen, and subsequently stored at ‑80°C until use. All participants signed the written informed consent and this study was approved by the Ethic Committee of The First Hospital of Tianshui (Gansu, China).
Table 1. Clinicopathological characteristics in NSCLC patients (n = 30).
Human NSCLC cell lines (H1299, 95D, H1650, SPC-A1, and A549) and the the normal immortalized bronchial epithelial cell line, BEAS-2B were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). H1299, 95D and H1650 cells were cultured in RPMI-1640 medium (Invitrogen; Thermo Fisher Scientifc, Inc., Waltham, MA, USA) with 10% fetal bovine serum (FBS, Gibco, NY, USA). A549 and BEAS-2B cell lines were cultivated in Dulbecco’s Modified Eagle’s Medium (DMEM, Gibco) containing 10% FBS (Gibco). All cell lines were maintained in a humidified atmosphere with 5% CO2 at 37 °C.
Cell transfection
The human miR-5195-3p mimics (miR-5195-3p) and negative control mimics (miR-NC) were synthesized by Shanghai GenePharma Co., Ltd. (Shanghai, China). Chemically modified small interfering RNA for MYO6 (siMYO6) and negative control siRNA (siNC) were obtained from RuiBoBio (Guangzhou, China). The open reading frame of MYO6 was cloned into the pcDNA3.1 vector (Invitrogen) to construct an MYO6 eukaryotic expression vector (MYO6/pcDNA3.1). The empty pcDNA3.1-vector was used as control.
95D and A549 cells were seeded in a 6‑well plate in a density of 5 × 105 cells per well and incubated at 37 °C overnight to reach 60‑70% confluence. For miR-5195-3p overexpression, 95D and A549 cells were transfected with 50 nM miR-5195-3p or miR-NC, respectively. For MYO6 silencing, 95D and A549 cells were transfected with siMYO6 or siNC, respectively. For the rescue experiments, MYO6 expressing plasmid or pcDNA3.1-vector was transfected into 95D cells transiently transfected with miR-5195-3p or miR-NC, respectively. All oligonucleotides were diluted in serum-free Opti-minimal essential medium (MEM) and cell transfection was performed using Lipofectamine® 2000 (Invitrogen, Thermo Fisher Scientific, Inc.) according to the manufacturer’s instructions. After 48 h transfection, transfected cells were used for the subsequent experiments.
Bioinformatics predication
Publicly available tools including TargetScan (http://www.targetscan.org/), miRanda (http://www.targetscan.org/) and PicTar (http://pictar.mdc-berlin.de/) were used to predict the candidate target genes of miR-5195-3p.
Dual-luciferase reporter assay
According to bioinformatics predictions, the wild-type of miR-5195-3p binding sequences on the 3ʹ-UTR region of MYO6 gene were synthesized by Ribobio Co. (Guangzhou, China). The miR-5195-3p binding sites within the MYO6 3ʹ-UTR were mutated to remove the binding sites. Next, the wild-type (WT) or mutant (MUT) 3ʹ-UTR of MYO6 was cloned into the pmirGLO dual-luciferase vector (Promega, WI, USA) between the Xho I and Sal I sites by Ribobio Co. (Guangzhou, China). For the luciferase reporter assay, 95D and A549 cells (1 × 105 cells per well) were seeded in 24-well plates and co-transfected with 300 ng of WT or MUT luciferase vector together with 30 nM of either miR-5195-3p or miR-NC. Then the luciferase activity was determined at 48 h after transfection using a Dual-Luciferase® Assay System (Promega) according to the manufacturer’s instructions.
RNA extraction and quantitative real-time PCR
Total RNA was extracted from tissue samples or cell lines using TRIzol Reagent (Invitrogen) and the cDNA was synthesized using a PrimeScript RT reagent kit (Takara, Dalian, China) following the manufacturer’s instructions. The stem-loop RT primers for miR-5195-3p and U6 were purchased from RiboBio Co. (Guangzhou, China). Gene-specific primers for MYO6 (forward: 5′-CAGAGCAACGTGCTCCAAAGTC’-3′ and reverse: 5′-GAAGCGTTGCTGTCGGTTCA-3′) and GAPDH (forward: 5′-GTGGACCTGACCTGCCGTCT-3′ and reverse: 5′-GGAGGAGTGGGTGTCGCTGT-3′) were synthesized by Shanghai Sangon Biotechnology Co. (Shanghai, China). Real-time PCR analysis was performed using an Applied Biosystems 7900HT Fast Real-Time PCR System instrument (Applied Biosystems). The comparative threshold cycle (CT) (2−ΔΔCt) method was used to obtain the relative fold changes in gene expression. The U6 small nuclear RNA and GAPDH were used as internal controls for the miR-5195-3p and MYO6 mRNA assays, respectively.
Western blot analysis
Transfected cells were harvested and lysed in cold RIPA buffer with a protease inhibitor (Beyotime, Shanghai, China). The supernatants were collected through centrifugation at 4 °C for 10 min at 12,000 × g. After protein quantification with BCA protein assay kit (Pierce, IL, USA), Equal amounts of protein were separated with 10% SDS-PAGE and transferred to a polyvinylidene difluoride (PVDF) membrane (Millipore, Hercules, CA, USA). Following 2 h block with 5% skimmed milk in Tris-buffered saline containing 0.1% Tween-20 (TBST), the membranes were incubated with primary antibodies against MYO6 (cat. no. sc-50,461; Santa Cruz Biotechnology, Inc.) and GAPDH (cat. no. 60,186–1-1g, Proteintech) overnight at 4 °C. After washing with TBST three times, the membrane was incubated with horseradish peroxidase-conjugated secondary antibody (cat. no. sc-2005; Santa Cruz Biotechnology, Inc.) for 2 h. Band signals were visualized by enhanced chemiluminescence kit (GE Healthcare Life Sciences, Shanghai, China).
CCK-8 proliferation assay
A CCK-8 assay (Dojindo Laboratories, Kumamoto, Japan) was performed to examine cell proliferation in NSCLC cells. Briefly, 95D or A549 cells were collected after 48 h transfection, and then cultured into 96-well plates (3 × 103 cell per well) for 24, 48, 72 and 96 h, respectively. Subsequently, 10 μL CCK-8 solution was added to each well, followed by incubation for 2 h at 37 °C. The optical density (OD) value at 450 nm was determined using a microplate reader (Thermo Fisher Scientific, Inc.). According to the OD value at each indicated time point, the cell proliferation curves were plotted. Each sample was prepared in triplicate and the experiment was conducted in three times.
Transwell assay
A migration assay was carried out to analyze the migratory capacity of transfected cells using a Transwell membrane (8 µm pore size; Corning-Costar). For the invasion assay, the Transwell membrane was coated with Matrigel® (BD Biosciences, NJ, USA). Briefly, transfected cells (1 × 106 cells) were re-suspended in serum-free medium and added to the upper chambers. Approximately 600 μL complete medium containing 10% FBS was added to the lower chambers as a chemoattractant. After 24 h incubation, the cells that had migrated to the lower chamber were fixed in 20 % methanol and stained with 0.1% crystal violet for 20 min at room temperature. After washing with PBS, five random fields were selected using a light microscope (magnification, × 200) and number of cells was counted to calculate the average migrated cell number was calculated.
Statistical analysis
Results were expressed as means ± standard deviation (SD). All statistical analysis was performed using GraphPad Prism software version 6.0 (GraphPad, Software, Inc., La Jolla, CA, USA). The correlation between miR-5195-3p and MYO6 expression in NSCLC tissues was determined using Pearson’s correlation coefficient by GraphPad Prism 6.0 software. Statistical differences were evaluated using Student’s t-test for two-group comparisons and one-way analysis of variance for more than two groups. The p-value of less than 0.05 was considered statistically significant.
Results
The expression of mir‑5195‑3p was significantly down-regulated in NSCLC tissues and cell lines
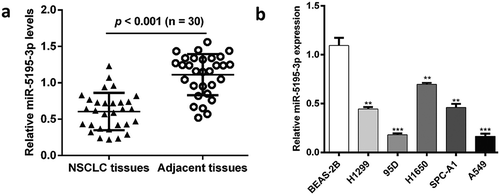
To explore the potential function of miR‑5195‑3p in NSCLC, we used quantitative real-time PCR to determine its expression in 30 paired human NSCLC tissues and the matched adjacent non-tumorous lung tissues. As show in , miR‑5195‑3p was significantly decreased in NSCLC tissues, as compared with non-tumorous lung tissues (p < 0.001). Additionally, significantly decreased miR‑5195‑‑3p expression in the five NSCLC cell lines, H1299, 95D, H1650, SPC-A1, and A549 was also observed, in comparison with BEAS-2B cells (, p < 0.01, p < 0.001). Among these five NSCLC cell lines, 95D and A549 cells expressed lower miR‑5195‑3p, which thus were selected for all subsequent experiments.
Figure 1. Low miR-5195-3p expression was found in NSCLC tissues and cell lines. (a) miR-5195-3p expression in 30 pairs of NSCLC tissues was determined using quantitative real-time PCR assays, compared with the adjacent tissues. (b) miR-5195-3p expression in NSCLC cell lines H1299, 95D, H1650, SPC-A1, and A549, and normal cell line BEAS-2B using quantitative real-time PCR assay. The data are presented as the mean ± standard deviation of three independent experiments. **p < 0.01, ***p < 0.001 vs. BEAS-2B; NSCLC, non-small cell lung cancer.
Mir-5195-3p inversely regulates MYO6 expression through direct binding to its 3ʹUTR
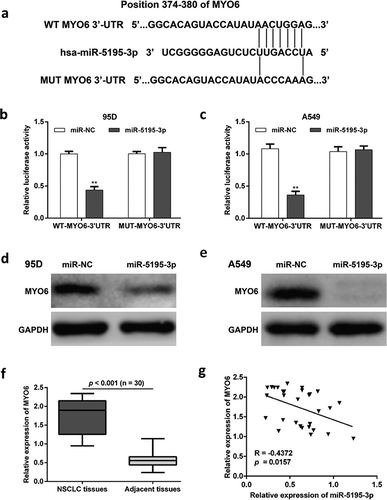
Since MYO6 has been reported to act as an oncogene in several tumors, I thus speculated that it might be regulated by a number of miRNAs. As expected, online bioinformatics software showed that there was a binding site of miR-5195-3p in the 3ʹ-UTR region of MYO6. The wild-type and mutant type of miR-5195-3p binding sequence is shown in . To confirm that MYO6 is a direct target gene of miR-5195-3p, the luciferase reporter assay was performed in 95D and A549 cells co-transfected with miR-NC or miR-5195-3p together with WT or MUT MYO6-3ʹ-UTR. The results showed indicated that overexpression of miR-5195-3p significantly decreased the relative luciferase activity of WT-MYO6-3ʹUTR, but did not alter that of MUT-MYO6-3ʹUTR in both 95D (, p < 0.01) and A549 cells (, p < 0.01). Moreover, western blot analysis was performed to confirm the regulatory effect of miR-5195-3p on MYO6 expression level in NSCLC cells. As shown in and , overexpression of miR-5195-3p obviously down-regulated the expression level of MYO6 protein in both 95D and A549 cells. In addition, quantitative real-time PCR revealed a significantly increased MYO6 expression levels in 30 pairs of NSCLC tissues compared with the matched adjacent lung tissues (). Pearson’s correlation analysis further demonstrated that there was significantly negative correlation between miR-5195-3p and MYO6 in NSCLC tissues (, p = 0.0157). Collectively, these results indicated that miR-5195-3p negatively regulated MYO6 expression by directly binding to its 3ʹUTR region in NSCLC cells.
Figure 2. MiR-5195-3p inversely regulates MYO6 expression through direct binding to its 3ʹUTR. (a) Complementary sequences between miR-5195-3p and the 3ʹ-UTR of MYO6 mRNA were obtained using publicly available algorithms. The mutated version of the MYO6 3ʹ-UTR is also shown. The relative luciferase activities were inhibited in (b) 95D and (c) A549 cells co-transfected with wild-type MYO6 3ʹUTR vector and miR-5195-3p, not with the mutant-type vector. Firefly luciferase activity was normalized to Renilla luciferase. Data are presented as the mean value ± SD from triplicate experiments. **p < 0.01 vs. miR-NC; Western blot analysis was performed to determine the protein level of MYO6 in (d) 95D and (e) A549 cells transfected with miR-5195-3p or miR-NC. (f) The relative expression levels of MYO6 mRNA in NSCLC tissues and adjacent tissues were detected by quantitative real time PCR. (g) The Pearson’s correlation analysis for the correlation between miR-5195-3p levels and MYO6 mRNA in NSCLC tissues.
Up-regulation of mir-5195-3p suppressed cell proliferation, migration and invasion in NSCLC cells
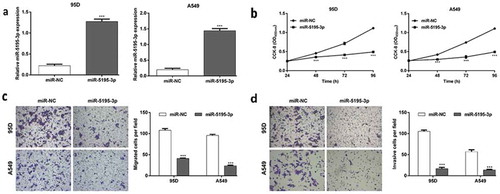
As the expression of miR-5195-3p was down-regulated in NSCLC tissues and cell lines, it might function as a tumor suppressor in NSCLC progression. To confirm this, a series of gain-of-function assays were performed to evaluate the function of miR-5195-3p in NSCLC cells. As shown in , quantitative real-time PCR firstly demonstrated that the expression of miR-5195-3p was significantly increased in both 95D and A549 cells after transfected with miR-5195-3p, compared with miR-NC transfection (p < 0.001). CCK-8 assay showed that overexpression of miR-5195-3p significantly inhibited cell proliferation in the 95D and A549 cells (, p < 0.001). Furthermore, the effects of miR-5195-3p overexpression on cell migration and invasion were evaluated using Transwell chamber assay. As shown in , the migratory cell numbers of miR-5195-3p group were significantly decreased compared with those of miR-NC group (95D: 41.0 ± 1.0 vs. 108.0 ± 4.0; A549: 23.7 ± 1.5 vs. 95.7 ± 2.5, p < 0.001). Similarly, the number of invasive cells of miR-5195-3p group was remarkably less than that in the miR-NC group (, 95D: 16.7 ± 3.2 vs. 105.3 ± 3.1; A549: 13.7 ± 1.5 vs. 57.0 ± 4.6, p < 0.001).
Figure 3. Functions of miR-5195-3p overexpression in the regulation of NSCLC cell proliferation, migration and invasion. 95D and A549 cells were transfected with miR-5195-3p or miR-NC, respectively. (a) Quantitative real time PCR was used to analyze the expression levels of miR-5195-3p. (b) Cell proliferation was measured by CCK-8 assay. (c) The cell migration and (d) invasion capability of 95D and A549 cells was determined using Transwell assays. Data are presented as the mean value ± SD from triplicate experiments. ***p < 0.001 vs. miR-NC.
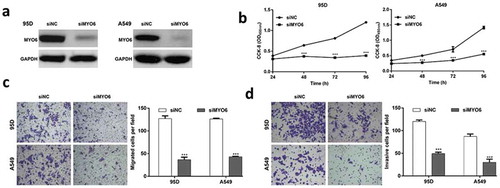
Knockdown of MYO6 inhibited cell proliferation, migration and invasion in NSCLC cells
To investigate the biological functions of MYO6 in NSCLC cells, endogenous MYO6 was knocked-down in 95D and A549 cells with a specific siMYO6. Western blotting confirmed MYO6 was significantly inhibited in 95D and A549 cells by siMYO6 (). Similar with miR-5195-3p overexpression, MYO6-knockdown in 95D and A549 cells inhibited cell growth and proliferation in vitro (, p < 0.001). Consistently, MYO6 knockdown significantly inhibited cell migration by 71.1% in 95D cells and 66.0% in A549 cells, respectively (, p < 0.001). The inhibition on cell invasion was 59.0% in 95D cells and 65.5% in A549 cells by MYO6 knockdown, respectively (, p < 0.001). These results implied that MYO6 knockdown attenuated the proliferation, migration and invasion of NSCLC cells, which was a similar effect exhibited by miR-5195-3p overexpression.
Figure 4. Functions of MYO6 knockdown in the regulation of NSCLC cell proliferation, migration and invasion. 95D and A549 cells were transfected with siMYO6 or siNC, respectively. (a) Western blot analysis was performed to analyze the protein level of MYO6. (b) Cell proliferation was evaluated by CCK-8 assay. (c) The cell migration and (d) invasion capability of 95D and A549 cells was measured using Transwell assays. Data are presented as the mean value ± SD from triplicate experiments. ***p < 0.001 vs. siNC.
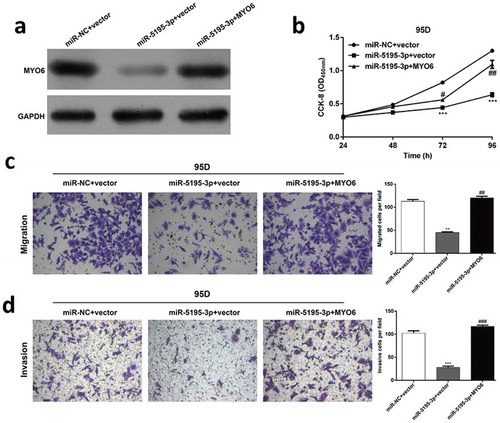
Overexpression of MYO6 reversed the inhibitory effects of mir-5195-3p on the proliferation, migration, and invasion of NSCLC cells
Next, the functional relevance of MYO6 targeting by miR-876-3p was investigated by determining whether MYO6 overexpression could rescue the inhibitory effects of miR-5195-3p on NSCLC cell proliferation, migration and invasion. 95D cells were co-transfected with miR-5195-3p and MYO6 overexpression plasmids. Western blot analysis was used to validate the MYO6 protein in the rescue experiments (). CCK-8 analysis suggested that the exogenous expression of MYO6 rescued the inhibitory effect of miR-5195-3p on cell proliferation in vitro (, p < 0.05, p < 0.01, p < 0.001). Transwell assays showed that the migration (, p < 0.01) and invasion ability (, p < 0.001) inhibited by miR-5195-3p was remarkably rescued by the MYO6 overexpression. These results suggested that miR-5195-3p influenced the growth and mobility of NSCLC cells by targeting MYO6.
Figure 5. MYO6 overexpression can partially rescue the effects of miR-5195-3p in NSCLC. Transfected 95D cells were divided into three groups, including miR-NC + vector, miR-5195-3p + vector, and miR-5195-3p + MYO6. (a) Western blot analysis was performed to analyze the protein level of MYO6. (b) Cell proliferation was measured by the CCK-8 assay. The CCK-8 assay was performed every 24 hours for 4 days. (c) The cell migration and (d) invasion capability of 95D. Data are presented as the mean value ± SD from triplicate experiments. **p < 0.01, ***p < 0.001 vs. miR-NC + vector, #p < 0.05, ##p < 0.01 vs. miR-5195-3p + vector.
Discussion
Cancer is generally considered a complex and non-site-specific disease to which many unknown etiologies contribute to tumor initiation and progression [Citation22]. The current work has led to the identification of miR-5195-3p, a low-expressed gene in NSCLC tissues and cell lines, prevents synthesis of MYO6 protein, permitting the inhibition of NSCLC cells proliferation, migration and invasion. Therefore, it is high likely that miR-5195-3p play a crucial role in suppression of NSCLC cells behavior and malignant transformation.
Previous studies proposed that more than 30% of all human genes are miRNA targets, and play a fundamental role in modulating [Citation23]. In fact, only several myosin family members are identified as target genes of miRNAs. Such as, myosin 1B provides a novel target of miR-363 to abrogate head and neck cancer cells migration [Citation24]. Fast myosin heavy chain isoforms are targeted by miR-23a to suppresses myogenic differentiation [Citation23]. In the present study, bioinformatics and luciferase reporter assay revealed that MYO6 is a novel direct target of miR-5195-3p. Moreover, MYO6 mRNA and protein levels are efficiently depressed by miR-5195-3p overexpression. Consistently, MYO6 has been identified as a target gene of miR-143 and miR-145 in gastric cancer, which is suppressed by both of them [Citation25]. In colorectal cancer, miR-145 could negatively regulate the expression of MYO6 as a target gene of miR-145 [Citation26]. Thus, MYO6 might play an important role in tumor progression and could be negatively regulated by various miRNAs.
Dissemination of tumor cells in patients with NSCLC is a difficult clinical challenge, and its underlying molecular mechanisms are still poor [Citation27]. In the present study, MYO6, a minus-end-directed actin motor, was observed to be high expressed in NSCLC tissues rather than adjacent tissues. According to the study from Lei et al, up-regulation of MYO6 has been found to be associated with larger tumor size and more frequent metastasis in gastric cancer patients [Citation25]. In addition, MYO6 was found to be significantly up-regulated in prostate cancer tissues and closely related with Gleason score [Citation28]. Further functional analysis showed that knockdown of MYO6 is responsible for inhibition of NSCLC cells proliferation, invasion and migration induced by overexpression of miR-5195-3p. In rat pheochromocytoma cells (PC12), Majewski et al. [Citation29] found that MYO6 deficiency not only impaired cells size and morphology, but also engaged in organization of actin cytoskeleton and Golgi apparatus. He also revealed that increased G0/G1 phase arrest is likely to be a major cause of reduced proliferation in PC12 cells [Citation29]. Besides, MYO6 is up-regulated in human breast cancer ZR-75–30 and MDA-MB-231 cells, and depletion of its expression also prevents cells proliferation [Citation30]. Here, MYO6 is identified as an oncogene in NSCLC, and suggesting that the gene may be implicated in regulation of NSCLC cells division.
Characterizing the action of MYO6 in the migratory of cells has aroused the people’s attention with the discovery that MYO6 is associated with cancer cells invasion [Citation31]. It has been shown that border-cell migration in the Drosophila ovary provides a good model for investigation the role of MYO6 in cancer cells invasion [Citation32,Citation33]. In fact, MYO6 plays a pivotal role in mechanical border cell migration that mediated by E-cadherin [Citation33]. Drosophila melanogaster MYO6 could stabilizes E-cadherin and Armadillo (Arm, Drosophila β-catenin) and develop a force of protrusive that enable to push the actin filaments outwards and drive cell-movement [Citation31]. In another previous study, cell spreading and migration were shown to be inhibited in high-grade ovarian cancer cells with MYO6 depletion [Citation32]. In the present study, observation that in NSCSL cells knockdown of MYO6 caused impairs of cell migration and invasion are most probably related to minus-end-directed motor properties in MYO6.
In conclusion, overexpression of miR-5195-3p can impede the proliferation, invasion and migration capabilities of NSCLC cells though negative post-transcriptional regulation of MYO6. Thus, this study may provide new clues for the development of therapeutics that prevent NSCLC dissemination.
Authors’ contributions
YQF designed, undertook, and performed the study. YQF performed the analysis and interpretation of data, and drafted and revised the manuscript.
Acknowledgments
The study was supported by The First Hospital of Tianshui (Gansu, China).
Disclosure statement
No potential conflict of interest was reported by the author.
References
- Molina JR, Yang P, Cassivi SD, et al. Non-small cell lung cancer: epidemiology, risk factors, treatment, and survivorship. Mayo Clin Proc. 2008 May;83(5):584–594. PubMed PMID: 18452692; PubMed Central PMCID: PMCPMC2718421.
- Huber RM. Is lung cancer in never-smokers a different disease?–back to the figures. J Thorac Oncol. 2007 Sep;2(9):787–788. PubMed PMID: 17805053.
- Mehta A, Dobersch S, Romero-Olmedo AJ, et al. Epigenetics in lung cancer diagnosis and therapy. Cancer Metastasis Rev. 2015 Jun;34(2):229–241. PubMed PMID: 25939322.
- Gridelli C, Rossi A, Carbone DP, et al. Non-small-cell lung cancer [Primer]. Nat Rev Dis Primers. 2015 05/21/online;1:15009.
- Negrini M, Calin GA. Breast cancer metastasis: a microRNA story. Breast Cancer Res. 2008;10(2):203. PubMed PMID: 18373886; PubMed Central PMCID: PMCPMC2397516.
- Iorio MV, Ferracin M, Liu CG, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005 Aug 15;65(16):7065–7070. PubMed PMID: 16103053.
- Asangani IA, Rasheed SA, Nikolova DA, et al. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008 Apr 3;27(15):2128–2136. PubMed PMID: 17968323.
- Kong W, He L, Richards EJ, et al. Upregulation of miRNA-155 promotes tumour angiogenesis by targeting VHL and is associated with poor prognosis and triple-negative breast cancer. Oncogene. 2014 Feb 6;33(6):679–689. PubMed PMID: 23353819; PubMed Central PMCID: PMCPMC3925335.
- Imam JS, Buddavarapu K, Lee-Chang JS, et al. MicroRNA-185 suppresses tumor growth and progression by targeting the six1 oncogene in human cancers. Oncogene. 2010 Sep 2;29(35):4971–4979. PubMed PMID: 20603620.
- Dong P, Kaneuchi M, Watari H, et al. MicroRNA-194 inhibits epithelial to mesenchymal transition of endometrial cancer cells by targeting oncogene BMI-1. Mol Cancer. 2011 Aug 18;10:99. . PubMed PMID: 21851624; PubMed Central PMCID: PMCPMC3173388.
- Jiang Z, Zhang Y, Cao R, et al. miR-5195-3p inhibits proliferation and invasion of human bladder cancer cells by directly targeting oncogene KLF5. Oncol Res. 2017 Aug 7;25(7):1081–1087. PubMed PMID: 28109084.
- Zhang Z, Tian H, Miao Y, et al. Upregulation of p72 enhances malignant migration and invasion of glioma cells by repressing beclin1 expression. Biochemistry (Mosc). 2016 Jun;81(6):574–582. PubMed PMID: 27301285.
- Seiler C, Ben-David O, Sidi S, et al. Myosin VI is required for structural integrity of the apical surface of sensory hair cells in zebrafish. Dev Biol. 2004 Aug 15;272(2):328–338. PubMed PMID: 15282151.
- Wells AL, Lin AW, Chen LQ, et al. Myosin VI is an actin-based motor that moves backwards. Nature. 1999 Sep 30;401(6752):505–508. PubMed PMID: 10519557.
- Masters TA, Buss F. Filopodia formation and endosome clustering induced by mutant plus-end-directed myosin VI. Proc Natl Acad Sci U S A. 2017 Feb 14;114(7):1595–1600. PubMed PMID: 28143933; PubMed Central PMCID: PMCPMC5320995.
- You N, Li J, Gong Z, et al. COMMD7 functions as molecular target in pancreatic ductal adenocarcinoma. Mol Carcinog. 2016;56(2):607–624.
- Mohiddin SA, Ahmed ZM, Griffith AJ, et al. Novel association of hypertrophic cardiomyopathy, sensorineural deafness, and a mutation in unconventional myosin VI (MYO6). J Med Genet. 2004 Apr;41(4):309–314. PubMed PMID: 15060111; PubMed Central PMCID: PMCPMC1735721.
- Avraham KB, Hasson T, Sobe T, et al. Characterization of unconventional MYO6, the human homologue of the gene responsible for deafness in Snell’s waltzer mice. Hum Mol Genet. 1997 Aug;6(8):1225–1231. PubMed PMID: 9259267.
- Dunn TA, Chen S, Faith DA, et al. A novel role of myosin VI in human prostate cancer. Am J Pathol. 2006 Nov;169(5):1843–1854. PubMed PMID: 17071605; PubMed Central PMCID: PMCPMC1780223.
- Gleason DF, Mellinger GT. Veterans administration cooperative urological research G. Prediction of prognosis for prostatic adenocarcinoma by combined histological grading and clinical staging. 1974. J Urol. 2002 Feb;167(2 Pt 2):953–958. discussion 959. PubMed PMID: 11905924.
- Kay FU, Kandathil A, Batra K, et al. Revisions to the tumor, node, metastasis staging of lung cancer (8(th) edition): rationale, radiologic findings and clinical implications. World J Radiol. 2017 Jun 28;9(6):269–279. PubMed PMID: 28717413; PubMed Central PMCID: PMC5491654.
- Amundadottir LT, Thorvaldsson S, Gudbjartsson DF, et al. Cancer as a complex phenotype: pattern of cancer distribution within and beyond the nuclear family. PLoS Med. 2004 Dec;1(3):e65. PubMed PMID: 15630470; PubMed Central PMCID: PMCPMC539051.
- Sood P, Krek A, Zavolan M, et al. Cell-type-specific signatures of microRNAs on target mRNA expression. Proc Natl Acad Sci U S A. 2006 Feb 21;103(8):2746–2751. PubMed PMID: 16477010; PubMed Central PMCID: PMCPMC1413820.
- Chapman BV, Wald AI, Akhtar P, et al. MicroRNA-363 targets myosin 1B to reduce cellular migration in head and neck cancer. BMC Cancer. 2015 Nov 6;15:861. . PubMed PMID: 26545583; PubMed Central PMCID: PMCPMC4635687.
- Lei C, Du F, Sun L, et al. miR-143 and miR-145 inhibit gastric cancer cell migration and metastasis by suppressing MYO6. Cell Death Dis. 2017 Oct 12;8(10):e3101. PubMed PMID: 29022908; PubMed Central PMCID: PMC5682659.
- Wei AW, Li LF. Long non-coding RNA SOX21-AS1 sponges miR-145 to promote the tumorigenesis of colorectal cancer by targeting MYO6. Biomed Pharmacother. 2017 Dec;96:953–959. PubMed PMID: 29217166.
- Kasinski AL, Kelnar K, Stahlhut C, et al. A combinatorial microRNA therapeutics approach to suppressing non-small cell lung cancer. Oncogene. 2015 Jul;34(27):3547–3555. PubMed PMID: 25174400; PubMed Central PMCID: PMCPMC4345154.
- Wang D, Zhu L, Liao M, et al. MYO6 knockdown inhibits the growth and induces the apoptosis of prostate cancer cells by decreasing the phosphorylation of ERK1/2 and PRAS40. Oncol Rep. 2016 Sep;36(3):1285–1292. PubMed PMID: 27431378.
- Majewski L, Sobczak M, Wasik A, et al. Myosin VI in PC12 cells plays important roles in cell migration and proliferation but not in catecholamine secretion. J Muscle Res Cell Motil. 2011 Dec;32(4–5):291–302. PubMed PMID: 22105702; PubMed Central PMCID: PMCPMC3230755.
- Wang H, Wang B, Zhu W, et al. Lentivirus-mediated knockdown of myosin VI inhibits cell proliferation of breast cancer cell. Cancer Biother Radiopharm. 2015 Oct;30(8):330–335. PubMed PMID: 26407123.
- Knudsen B. Migrating with myosin VI. Am J Pathol. 2006 Nov;169(5):1523–1526. PubMed PMID: 17071577; PubMed Central PMCID: PMCPMC1780205.
- Yoshida H, Cheng W, Hung J, et al. Lessons from border cell migration in the Drosophila ovary: A role for myosin VI in dissemination of human ovarian cancer. Proc Natl Acad Sci U S A. 2004 May 25;101(21):8144–8149. PubMed PMID: 15146066; PubMed Central PMCID: PMCPMC419571.
- Geisbrecht ER, Montell DJ. Myosin VI is required for E-cadherin-mediated border cell migration. Nat Cell Biol. 2002 Aug;4(8):616–620. PubMed PMID: 12134162.
