ABSTRACT
Recently, miR-221-3p expression has been reported to be down-regulated in medulloblastoma (MB), but its functional effects remains unclear. In this study, quantitative real-time PCR (qRT-PCR) revealed significantly decreased miR-221-3p in MB cell lines. Transfection of miR-221-3p mimics reduced, or inhibitor increased cell proliferation in MB cells using MTT assay. Flow cytometry analysis indicated miR-221-3p overexpression promoted, while knockdown alleviated G0/G1 arrest and apoptosis. Luciferase reporter assay confirmed miR-221-3p directly targets the EIF5A2 gene. Moreover, restoration of EIF5A2 in the miR-221-3p-overexpressing DAOY cells significantly alleviated the suppressive effects of miR-221-3p on cell proliferation, cell cycle and apoptosis. Furthermore, miR-221-3p overexpression decreased CDK4, Cyclin D1 and Bcl-2 and increased Bad expression, which was reversed by EIF5A2 overexpression. These results uncovered the tumor suppressive role of miR-221-3p in MB cell proliferation at least in part via targeting EIF5A2, suggesting that miR-221-3p might be a potential candidate target for diagnosis and therapeutics of MB.
Graphical Abstract
MiR-221-3p overexpression significantly suppressed medulloblastoma cell proliferation, induced cell cycle G0/G1 phase arrest and apoptosis by directly targeting the EIF5A2 gene.
Medulloblastoma (MB) is a highly aggressive and treatment-refractory paediatric cerebellar brain tumor [Citation1]. With the development of combined surgery, radiation, and chemotherapy, the survival rates in MB patients have been improved in the last few decades [Citation2]. However, these traditional methods have posed a considerable toxicity burden to developing child, thus 25–30% of patients would die from the disease. In addition, most survivors may suffer from the side effects such as neurologic, endocrinologic, and social sequelae [Citation3,Citation4]. MB consists of four types of subgroups: WNT, sonic hedgehog (SHH), Group 3, and Group 4 [Citation5]. Although MB is usually considered to be a single malignant, it is appeared to have a high degree of heterogeneity [Citation3]. Molecular research on the biological function of MB cells is pivotal to improve clinical trial design and develop the therapeutic targets for MB.
MicroRNAs (miRNAs), are ~20 nt in length, non-coding single stranded RNA molecules that play a critical role in post-transcriptional regulatory programs underlying almost all fundamental biological processes, including cellular behavior, organismal metabolism and development [Citation6,Citation7]. Investigation on miRNA-mRNA networks has been considerably expanded since the identification of the first miRNA during the past two decades [Citation8]. Numerous mRNAs have been proposed as targets of each miRNA as estimated by miRNAs target genome-wide identification and computational predication [Citation7]. Dysregulation of miRNAs are generally considered to be causal in diverse types of cancer, where miRNAs functions as oncomiRs or anti-oncomiRs [Citation9]. Recently, miRNA family gene miR-221-3p was identified as an anti-oncogene in ovarian cancer and triple negative breast cancer that linked to better prognosis by suppressing ARF4 and PARP1 [Citation10,Citation11]. On the contrary, miR-221-3p was positively correlated with tumorigenesis and metastasis of cervical cancer and gastric carcinoma by targeting THBS2 and PTEN [Citation12,Citation13]. Li et al. [Citation14] suggest that circulating miR-221-3p may serve as a valuable predictive biomarker in pancreatic cancer. Importantly, a previous study revealed low expression of miR-221 in fixed-formalin paraffin-embedded (FFPE) brain tissues of MB by using quantitative real-time PCR, indicating that miR-221 may be crucial in the pathogenesis of MB [Citation15]. However, to our knowledge, there is no additional information available on the biological role of miR-221-3p in MB.
Eukaryotic translation initiation factor (EIF) 5A2 belongs to a member of EIF family, which is known as a small universally conserved acidic protein [Citation16]. EIF5A2 shares up to 83% amino acid pairwise identity with EIF5A1 as a highly conserved protein involved in multiple cellular processes [Citation17]. It has been recently found to be mapped to chromosome 3q25-q27, an unstable and tumor associated gene regions [Citation18]. Accumulating evidence indicates that EIF5A2 is rarely expressed in normal tissues, whereas highly expressed in ovarian cancer [Citation19], bladder cancer [Citation20], and colorectal cancer [Citation21]. In a previous study, another EIF family member EIF4E was found to form a feedback loop with Mnk2 and associated with MB etiology as well as molecular pathogenesis [Citation22]. Therefore, we speculated that EIF5A2 may also have an important role in the pathogenesis of MB.
In the present study, we first determined the expression of miR-221-3p in human MB cell lines and its biological function in MB cells. Next, we identified EIF5A2 as a possible downstream target and determined the biologic effects of EIF5A2 upregulation on MB cells. Our results may help understand the biological role of miR-221-3p and define new therapeutic strategies for treatment of MB.
Materials and methods
MB cell line and culture conditions
Human MB cell lines (D341: No. HTB-185; D283 Med: No. HTB-187) were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA). Another MB cell line, DAOY was obtained from the cell bank of the Chinese Academy of Sciences (Shanghai, China). D283 Med and DAOY cells were cultured in α-minimum essential medium (α-MEM; HyClone, Logan City, UT, USA) supplemented with 10% fetal bovine serum (FBS, Gibco) and 1% penicillin/streptomycin. The D341 cell line was cultured in DMEM with 20% FBS with 1% penicillin/streptomycin. All cell lines were maintained in a humidified incubator containing 5% CO2 at 37 °C.
Transfection studies
The miR-221-3p mimics, mimic negative control (miR-NC), miR-221-3p inhibitor and scramble control (anti-miR) were purchased from RiboBio Co., Ltd. (Guangzhou, China). The EIF5A2 overexpressing pcDNA3.1-EIF5A2 recombinant plasmid and empty pcDNA3.1 plasmid were synthesized from Genepharma Co., Ltd (Shanghai, China). Transfection was performed in D283 Med and DAOY cells in a final concentration of 50 nM of mimics or 100 nM of inhibitor and their corresponding NC or in a final concentration of the EIF5A2 expression plasmids or the empty vector was 10 µg using Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA). The transfected cells were harvested after 48 h for the following analysis.
RNA extraction and quantitative real-time PCR (qRT-PCR)
Total RNA from the cultured cells was extracted using the TRIzol reagent (Life Technologies Corporation, Carlsbad, CA, USA) according to the manufacturer’s instructions. For miR-221-3p expression, first-strand cDNA was first synthesized from 1 µg of RNA using Ncode™ miRNA First-Strand cDNA Synthesis kit (Invitrogen; Thermo Fisher Scientific, Inc.). Real-time PCR was carried out on a Bio-Rad CFX384 PCR instrument (Bio-Rad Laboratories, Inc., Hercules, CA, USA) with the amplification primers (forward: 5′-GCTACATTGTCTGCTGGGTTTC-3′; reverse: 5′-GAGACTGCGGATGTATAGAACTTGA-3′) using U6 small nuclear RNA (forward: 5′-GCTTCGGCAGCACATATACTAAAAT-3′; reverse: 5′-GAGACTGCGGATGTATAGAACTTGA-3′) as the internal reference.
To detect the EIF5A2 mRNA expression, 500 ng of RNA was reversed transcribed into cDNA using the Prime-Script RT reagent kit (TaKaRa, Dalian, China). Real-time PCR was carried out on a Bio-Rad CFX384 PCR instrument (Bio-Rad Laboratories, Inc., Hercules, CA, USA) with the amplification primers (forward: 5′-GGACGACCATGCAAAATAGTGG-3′; reverse: 5′-TGCCCGTGAAAATATCAATTCCA-3′) using GAPDH (primer: 5′-AGCCTTCTCCATGGTGGTGAA-3′; reverse: 5′-ATCACCATCTTCCAGGAGCGA-3′) as an internal control. Each sample was prepared in triplicate and determined at least three times. The 2–ΔΔCt method was used for data analysis.
Cell proliferation assay
MTT assay was used for the determination of MB cell proliferation. Briefly, transfected cells were seeded into 96-well plates at a density of 2.5 × 103 cells/well and incubated overnight. At the indicated culture time (1, 2, 3, 4 and 5 day, respectively), 20 µl of MTT (5 mg/mL, Sigma, cat.n.M2128) was added to each well, and the cells were incubated at 37 °C for another 2 h. Then, the supernatant was removed and 100 µl DMSO was added to terminate the reaction. Finally, the optical density (OD) was determined at a wavelength of 595 nm using an ELISA reader (BD Biosciences, Franklin Lake, NJ, USA).
Cell cycle analysis
Cell cycle phase distribution of MB cells was analyzed using flow cytometry. Briefly, transfected cells were reseeded in six-well plates at a density of 3 × 105 cells per well and cultured overnight for adhesion. Then cells were collected and fixed in ice-cold 70% ethanol overnight at 4 °C. After washing with PBS solution, the cells were incubated with 10 mg/mL RNase and 1 mg/mL propidium iodide (PI, BD Biosciences) at room temperature for 30 min. The distribution of cells in G0/G1, S and G2/M phase was analyzed with ModFit version 4.0 software (Verity Software House, Topsham, ME, USA) using FACScan flow cytometer (BD Biosciences, San Diego, CA, USA).
Cell apoptosis analysis
Cell apoptosis of MB cells was analyzed using Annexin V-FITC/PI Apoptosis Detection kit (BD Biosciences). Briefly, transfected cells at a density of 3 × 105 cells per well reseeded in six-well plates. Then they were resuspended in 200 μL Annexin V binding buffer, followed by incubation with FITC-conjugated Annexin V (5 µL) and PI (5 µL) for 15 min in the dark. Stained cells were analyzed using FACScan flow cytometer equipped with FACSDiva software (BD Biosciences, San Diego, CA, USA). The overall apoptotic cells, including early (Annexin V+/PI-) and late apoptotic (Annexin V+/PI+) cells were analyzed and statistically calculated.
Bioinformatics and luciferase reporter assay
The potential target genes of miR-221-3p were predicted by using online miRNA target prediction algorithms: TargetScan (http://www.targetscan.org/vert_71/), PicTar4 (http://pictar.mdc-berlin.de) and miRanda (www.microrna.org). The nucleotide sequence of the 3′ untranslated region (UTR) of EIF5A2 was identified as a potential target gene. Subsequently, a luciferase reporter construct was constructed to validate their relationship. Briefly, the wild type (Wt) EIF5A2 3ʹUTR containing putative miR-221-3p binding sites and the mutant (Mut) EIF5A2 3′UTR were inserted between the XhoI and NotI sites of the luciferase reporter vector psiCHECK-2 (Promega, Madison, WI, USA). Wild-type and mutant luciferase reporter plasmids were validated by DNA sequencing. Co-transfection of 2 μg reporter plasmid with 50 nmol/L miRNA into HEK293T cells was accomplished using Lipofectamine 2000. Forty-eight hours after transfection, luciferase activity was measured using the Dual-luciferase Reporter Assay System (Promega, United States) according to the manufacturer’s instructions. Three independent experiments were performed in triplicate.
Western blot analysis of protein
Whole cell lysates were prepared using ice-cold RIPA buffer containing protease and phosphatase (Beyotime Institute of Biotechnology). Equal amounts of protein were separated by a 12% SDS-PAGE and transferred to polyvinylidene fluoride (PVDF) membranes (Millipore, Billerica, MA, USA). After blocking with 5% milk in TBST for 2 h at room temperature, the membranes were incubated with the appropriate primary antibodies against EIF5A2 (ab126735, Abcam, Cambridge, UK), CDK4 (#2906, Cell Signaling Technology, MA, USA), Cyclin D1 (#60186–1-1 g, Proteintech Group, Inc. IL, USA), Bcl-2 (ab32124, Abcam), Bad (ab32445, Abcam) and GAPDH (#10494–1-AP, Proteintech Group, Inc.) overnight at 4 °C followed by appropriate secondary antibodies (1:5000) conjugated to horseradish peroxidase. The protein bands were visualized using ECL detection reagents (Pierce, USA). The relative protein expression levels were normalized to GAPDH.
Statistical analysis
All data are expressed as the means ± SD of at least three independent experiments. Statistical analysis was performed by GraphPad Prism software (GraphPad Software Inc., La Jolla, CA, USA). Differences between two groups were compared using a Student’s t-test and comparisons amongst three or more groups were made by analysis of variance. Differences were considered to be statistically significant when p < 0.05.
Results
MiR-221-3p overexpression significantly suppressed cell proliferation of MB cells
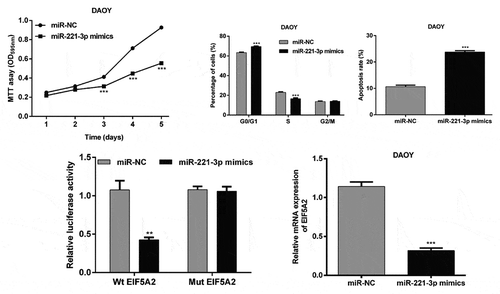
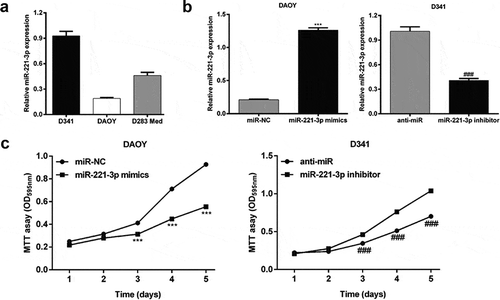
To explore the function role of miR-221-3p in MB in vitro, we firstly analyzed its expression in three MB cell lines, including DAOY, D341 and D283 Med using qRT-PCR. We found miR-221-3p presented the lowest expression in DAOY cells, but the highest expression in D341 cells ()). Then, DAOY and D341 cells were transfected with miR-221-3p mimics or inhibitor to overexpress or repress the expression of miR-221-3p, respectively. The results from qRT-PCR revealed miR-221-3p up-regulation in miR-221-3p mimics group of DAOY cells (5.1-fold) and down-regulation in miR-221-3p inhibitor group of D341 cells (59.6%), compared with the respective negative controls (), p < 0.001). In the MTT assay, stable miR-221-3p overexpression led to a significant reduction in cell proliferation ability of DAOY cells, compared with the respective control cells transfected with empty vector, at 3, 4 or 5 d time points. In contrast, down-regulation of miR-221-3p remarkably increased proliferation in D341 cells (), p < 0.001). These results indicate that miR-221-3p might play a suppressive role in regulating the MB cell proliferation.
Figure 1. MiR-221-3p affected medulloblastoma cell proliferation.
(a) Quantitative real-time PCR was used to determine the expression of miR-221-3p in medulloblastoma cell lines (D341, DAOY and D283 Med). (B) DAOY and D341 cells were transfected with miR-221-3p mimics or inhibitor to overexpress or repress the expression of miR-221-3p, respectively. Quantitative real-time PCR was used to determine the expression of miR-221-3p in DAOY and D341 cells. (c) Cell proliferation capacity of DAOY and D341 cells was measured by the MTT assay. All data were expressed as the mean ± standard deviation of at least three experiments. ***p< 0.001, as compared with miR-NC; ###p< 0.001, as compared with anti-miR
MiR-221-3p overexpression could promote cell cycle arrest and apoptosis of MB cells
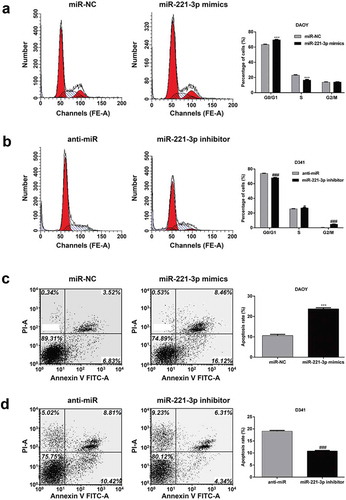
Furthermore, we used flow cytometry to assess the effects of miR-221-3p expression on cell cycle progression and apoptosis in MB cells. As shown in ), the percentage of DAOY cells at G1/G0 phase was significantly increased, and that in S phase was decreased in miR-221-3p mimics transfected cells, compared with those transfected with blank vector miR-NC (p < 0.001). In contrast, miR-221-3p knockdown significantly elevated the percentage of D341 cells at G1/G0 phase (p < 0.001), accordingly reduced the percentage of D341 cells at S (p < 0.05) and G2/M (p < 0.001) phase ()). These results demonstrated that miR-221-3p suppresses the cell cycle at G0/G1 phase in MB cells. In addition, flow cytometry further revealed a significant increase in the percentage of apoptotic DAOY cells in the miR-221-3p mimics transfection group compared with the miR-NC group (), p < 0.001), while a significant decrease in the percentage of apoptotic D341 cells in the miR-221-3p inhibitor transfection group compared with the anti-miR group (), p < 0.001). These results demonstrated that miR-221-3p promoted cell apoptosis in MB cells, which might contribute to the growth-inhibitory properties of miR-221-3p.
Figure 2. MiR-221-3p affected medulloblastoma cell cycle progression and apoptosis.
DAOY and D341 cells were transfected with miR-221-3p mimics or inhibitor to overexpress or repress the expression of miR-221-3p, respectively. Flow cytometry with PI staining were performed to measure the effect of miR-221-3p on cell cycle of (a) DAOY and (b) D341 cells. Representative flow cytometric histograms of each group showing the distribution of cell cycle were presented in left panel. Flow cytometry with Annexin V/PI staining were performed to measure the effect of miR-221-3p on cell apoptosis of (c) DAOY and (d) D341 cells. Representative flow cytometric histograms of each group showing the early apoptosis and late apoptosis were presented in left panel. All data were expressed as the mean ± standard deviation of at least three experiments. ***p< 0.001, as compared with miR-NC; #p< 0.05, ###p< 0.001, as compared with anti-miR
MiR-221-3p directly targets EIF5A2 in MB cells
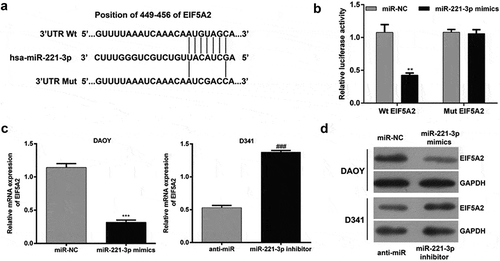
To elaborate the molecular mechanisms by which miR-221-3p regulating cell proliferation, cell cycle and apoptosis, we searched for candidate target genes of miR-221-3p that might be involved in the pathogenesis of MB. The bioinformatic analysis was performed to predict EIF5A2 as a putative target for miR-221-3p, which has been displayed in ). To verify this prediction, luciferase reporter constructs carrying the 3′ UTR miR-221-3p potential-binding site or corresponding mutant-binding site of EIF5A2 were constructed and co-transfected with miR-221-3p mimics or miR-NC into HEK293T cells. As shown in ), the luciferase activity was notably decreased after co-transfection with WT constructive luciferase reporter plasmid harboring the EIF5A2 3ʹUTR and miR-221-3p mimics (p < 0.01). However, miR-221-3p did not have an effect on the luciferase activity of the Mut 3ʹUTR-EIF5A2 luciferase reporter. Moreover, the overexpression of miR-221-3p led to a significant decrease in DAOY cells, while down-regulation of miR-221-3p caused a significant increase in D341 cells in EIF5A2 expression at the mRNA levels (), p < 0.001). Consistently, the results of western blot analysis also showed the expression of EIF5A2 protein levels was obviously down-regulated after miR-221-3p overexpression in DAOY cells, but up-regulated after miR-221-3p knockdown in D341 cells ()). These findings demonstrated that miR-221-3p might negatively regulate the expression of EIF5A2 by directly targeting the 449–456 nucleotide of its 3′-UTR.
Figure 3. EIF5A2 is a target gene of miR-221-3p in medulloblastoma.
(a) Graph indicating the wild-type and mutant binding site of miR-221-3p to EIF5A2, predicted by bioinformatics. (b) Relative luciferase activity of 293T cells co-transfected with miR-221-3p mimic and EIF5A2 3ʹUTR-luciferase reporter vector that contained a wild-type sequence (EIF5A2-3ʹUTR-wt), or in a vector that contained a mutant sequence (EIF5A2-3ʹUTR-mut) within the miR-221-3p-binding site. DAOY and D341 cells were transfected with miR-221-3p mimics or inhibitor to overexpress or repress the expression of miR-221-3p, respectively. (c) Quantitative real-time PCR and (d) western blot analysis of EIF5A2 expression in DAOY and D341 cells after 48 h transfection. All data were expressed as the mean ± standard deviation of at least three experiments. **p< 0.01, ***p< 0.001, as compared with miR-NC; ###p< 0.001, as compared with anti-miR
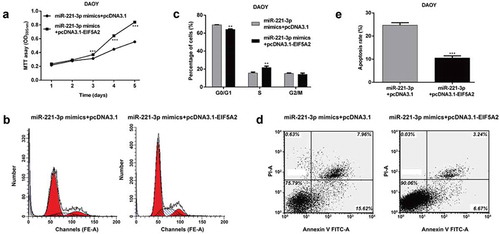
EIF5A2 is involved in miR-221-3p-mediated MB cell proliferation, cell cycle and apoptosis
Previous reports have shown that EIF5A2 is frequently up-regulated in tumor tissues and promotes tumor cell proliferation, and our results suggest that miR-221-3p could suppress MB cell proliferation and also down-regulated the expression of EIF5A2 by directly binding to its 3′ UTR. Therefore, we hypothesized that the EIF5A2 overexpression might directly mediate miR-221-3p-medicated cellular behavior in MB. Based on this speculation, we forced EIF5A2 expression in DAOY cells stably expressing miR-221-3p by transfecting with pcDNA-3.1-EIF5A2. MTT assay showed that the ectopic EIF5A2 expression in the miR-221-3p overexpressing cells attenuated the inhibitory effect of miR-221-3p on DAOY proliferation (), p < 0.001). Similarly, the induced cell cycle G0/G1 phase arrest () and (c), p < 0.01) and apoptosis () and (e), p < 0.001) by miR-221-3p overexpression was significantly reversed by overexpression of EIF5A2. Taken together, these results suggested that EIF5A2 was a direct functional downstream target of miR-221-3p in MB cells.
Figure 4. EIF5A2 is involved in miR-221-3p-mediated medulloblastoma cell proliferation, cell cycle and apoptosis.
DAOY cells were transfected with miR-221-3p mimics + pcDNA3.1 or miR-221-3p mimics + pcDNA3.1- EIF5A2 plasmid, respectively. The cell proliferation, cell cycle distribution and apoptosis were analyzed in DAOY cells using (a) MTT, (b and c) Flow cytometry with PI staining or (d and e) Flow cytometry with Annexin V-FITC/PI double staining, respectively. All data were expressed as the mean ± standard deviation of at least three experiments. **p< 0.01, ***p< 0.001, as compared with miR-221-3p mimics + pcDNA3.1
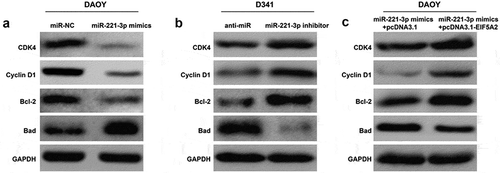
MiR-221-3p was involved in regulating cell cycle and apoptotic markers by targeting EIF5A2
To further elucidate the molecular mechanism underlying the regulation of miR-221-3p in cell cycle and apoptosis of MB cells, we next examined the influence of miR-221-3p by measuring the expression alterations of some cell cycle regulators and apoptotic markers. The results showed that transfection of miR-221-3p mimics led to decreased expression of CDK4, Cyclin D1 and Bcl-2 and increased Bad expression in DAOY cells ()), whereas silencing miR-221-3p elevated CDK4, Cyclin D1 and Bcl-2 expression, and reduced Bad expression in D341 cells ()). In addition, the attenuated expression of CDK4, Cyclin D1 and Bcl-2 and the up-regulated expression of Bad by miR-221-3p overexpression were reversed by the overexpression of EIF5A2 in DAOY cells ()). These results suggest that miR-221-3p significantly affected cell cycle G0/G1 phase and apoptotic markers by directly targeting EIF5A2 and eventually inhibits the cell proliferation in MB cells.
Figure 5. The effects of miR-221-3p on cell cycle regulators and apoptotic markers were mediated by EIF5A2 in medulloblastoma.
DAOY and D341 cells were transfected with miR-221-3p mimics or inhibitor to overexpress or repress the expression of miR-221-3p, respectively. DAOY cells were transfected with miR-221-3p mimics + pcDNA3.1 or miR-221-3p mimics + pcDNA3.1- EIF5A2 plasmid, respectively. Western blotting analysis of the expression of CDK4, Cyclin D1, Bcl-2 and Bad in (a) miR-221-3p overexpressing DAOY cells, (b) miR-221-3p knockdown D341 cells, and (c) overexpressing EIF5A2 DAOY cells after miR-221-3p mimics transfection.
Discussion
Recently, miRNAs have gained wide scholarly attention, likely due to their potential roles as post-transcriptional modulators of gene expression in eukaryotes [Citation23]. About 50% members of miRNA family are located in regions, frequently noted for chromosomal instability in human cancer, and involved in the pathogenesis of tumor through aberrant expression patterns [Citation24]. Among these dysregulated miRNAs, miR-221-3p is reported to be expressed at low levels in FFPE tissues of MB, but its functional role and the underlying molecular mechanisms in the malignant biological behavior of MB cells are still unclear. Therefore, the in vitro experiments were performed in MB cells to investigate the function role of miR-221-3p. By selecting several MB cell lines, we first found miR-221-3p exhibited the relative lower expression in DAOY cells, while higher expression in D341 cells.
Until now, recurrence and metastasis of cancer mainly attributed to the uncontrolled and usually rapid cellular growth [Citation25]. Our findings suggest that miR-221-3p-overexpression significantly suppressed cell proliferation capacity in DAOY cells while knockdown of miR-221-3p showed the opposite effect in D341 cells. To further investigate the molecular mechanisms regarding how miR-221-3p modulates MB cells proliferation, we analyzed cell cycle distribution and apoptosis rate. As a result, upregulation of miR-221-3p leaded to increased percentage of DAOY cells in G0/G1 phase and induced apoptosis. On the contrary, downregulation of miR-221-3p reversed that effect. These results indicating that miR-221-3p attenuated proliferation of MB cells might through blocking cell cycle progression and accelerating apoptosis.
It is well known that temporal activation of various cyclin-dependent kinases (CDK)/cyclins complexes, the core components of the cell cycle engine, are drivers of cell cycle events such as DNA replication, spindle formation, and chromosome segregation [Citation26]. CDK4/cyclin D and CDK6/cyclin D kinases are involved in ensuring the well-delineated transitions from G0/G1 onward into S phase by phosphorylation of the Retinoblastoma tumor suppressor protein (Rb) [Citation27]. Progression from S to G2 phase requires the activation of the CDK1/cyclin A and CDK2/cyclin E complexes, and CDK1/cyclin B complexes are sufficient for triggering mitosis and meiosis [Citation28]. Here, western blot analysis revealed that miR-221-3p-mimics drastically decreased, while miR-221-3p-inhibitor increased the levels of the CDK4 and cyclin D1 proteins, suggesting that miR-221-3p induces cells cycle arrest at G0/G1 phase in MB cells through the inhibition of CDK4/cyclin D1 complexes.
Apoptosis is considered as another complex biological process involved in cell survival and homeostasis [Citation29]. Anti-apoptotic and pro-apoptotic proteins are important controllers of apoptosis, their imbalance have been implicated in human cancer [Citation30]. Bcl-2, a key mediator of the mitochondrial and intrinsic apoptotic pathways, is amplified in almost all types of human malignancies, such as MB [Citation18], leukemia [Citation31], and lung cancer [Citation32]. It is interesting that the Bcl-2 proteins interact with pro-apoptotic Bcl-2 family members Bad and Bax to prevent apoptosis [Citation33]. In DAOY cells with miR-221-3p mimics transfection, we observed decreased Bcl-2 levels, but increased Bad levels, indicating anti-apoptotic and apoptotic homodimers imbalance, thus were responsible for apoptosis.
Besides, bioinformatics and luciferase reporter assays demonstrated that miR-221-3p targeted the 3ʹUTR of EIF5A2 in MB cells. To our knowledge, this is the first study showing evidence that EIF5A2 strongly associates with MB progression and development. EIF5A2 is a well described oncogene in several types of human cancer [Citation34]. Functionally, suppression of EIF5A2 in gastric cancer cells led to significant decreases in cell proliferation [Citation35]. Our further results indicated that ectopic expression of EIF5A2 reversed the inhibitory effect of miR-221-3p on MB cell proliferation, cell cycle progression and apoptosis, as well as molecular mechanisms. We thus considered that miR-221-3p might function as a tumor suppressor involved in the pathogenesis of MB via directly targeting EIF5A2.
In the present study, we present some evidences that miR-221-3p decreased MB cells proliferation though induction of G0/G1 arrest and apoptosis by down-regulating EIF5A2. Our studies may shed a light on the importance of miR-221-3p in the exploration of therapeutic target for MB.
Authors contributions
The study was designed by CHJ. The experiment was done by YY and WX. The data was acquired and analyzed by YY and WX. The manuscript was written by YY and WX. All authors read and approved the final manuscript
Acknowledgments
The study was supported by The Second People’s Hospital of Jingzhou.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Northcott PA, Lee C, Zichner T, et al. Enhancer hijacking activates GFI1 family oncogenes in medulloblastoma. Nature. 2014 Jul 24;511(7510):428–434. PubMed PMID: 25043047; PubMed Central PMCID: PMCPMC4201514.
- Srinivasan VM, Ghali MG, North RY, et al. Modern management of medulloblastoma: molecular classification, outcomes, and the role of surgery. Surg Neurol Int. 2016;7(Suppl 44):S1135–S1141. PubMed PMID: 28194300; PubMed Central PMCID: PMCPMC5299153.
- Wang J, Garancher A, Ramaswamy V, et al. Medulloblastoma: from molecular subgroups to molecular targeted therapies. Annu Rev Neurosci. 2018 Apr 11;41:207–232. PubMed PMID: 29641939.
- Northcott PA, Korshunov A, Witt H, et al. Medulloblastoma comprises four distinct molecular variants. J Clin Oncol. 2011 Apr 10;29(11):1408–1414. PubMed PMID: 20823417; PubMed Central PMCID: PMCPMC4874239.
- Kool M, Korshunov A, Remke M, et al. Molecular subgroups of medulloblastoma: an international meta-analysis of transcriptome, genetic aberrations, and clinical data of WNT, SHH, group 3, and group 4 medulloblastomas. Acta Neuropathol. 2012 Apr;123(4):473–484. PubMed PMID: 22358457; PubMed Central PMCID: PMCPMC3306778.
- Bandara KV, Michael MZ, Gleadle JM. MicroRNA Biogenesis in Hypoxia. Microrna. 2017;6(2):80–96. PubMed PMID: 28294076.
- Jonas S, Izaurralde E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat Rev Genet. 2015 Jul;16(7):421–433. PubMed PMID: 26077373.
- Cohen JL, Ata AE, Jackson NL, et al. Differential stress induced c-Fos expression and identification of region-specific miRNA-mRNA networks in the dorsal raphe and amygdala of high-responder/low-responder rats. Behav Brain Res. 2017 Feb 15;319:110–123. PubMed PMID: 27865919; PubMed Central PMCID: PMCPMC5183530.
- Xu G, Shao G, Pan Q, et al. MicroRNA-9 regulates non-small cell lung cancer cell invasion and migration by targeting eukaryotic translation initiation factor 5A2. Am J Transl Res. 2017;9(2):478–488. PubMed PMID: 28337276; PubMed Central PMCID: PMCPMC5340683.
- Deng L, Lei Q, Wang Y, et al. Downregulation of miR-221-3p and upregulation of its target gene PARP1 are prognostic biomarkers for triple negative breast cancer patients and associated with poor prognosis. Oncotarget. 2017 Dec 12;8(65):108712–108725. PubMed PMID: 29312562; PubMed Central PMCID: PMC5752475.
- Wu Q, Ren X, Zhang Y, et al. MiR-221-3p targets ARF4 and inhibits the proliferation and migration of epithelial ovarian cancer cells. Biochem Biophys Res Commun. 2018 Mar 18;497(4):1162–1170. PubMed PMID: 28057486.
- Wei WF, Zhou CF, Wu XG, et al. MicroRNA-221-3p, a TWIST2 target, promotes cervical cancer metastasis by directly targeting THBS2. Cell Death Dis. 2017 Dec 14;8(12):3220. 10.1038/s41419-017-0077-5. PubMed PMID: 29242498; PubMed Central PMCID: PMCPMC5870596.
- Shi J, Zhang Y, Jin N, et al. MicroRNA-221-3p plays an oncogenic role in gastric carcinoma by inhibiting PTEN expression. Oncol Res. 2017 Apr 14;25(4):523–536. PubMed PMID: 27712596.
- Li F, Xu JW, Wang L, et al. MicroRNA-221-3p is up-regulated and serves as a potential biomarker in pancreatic cancer. Artif Cells Nanomed Biotechnol. 2018 May;46(3):482–487. PubMed PMID: 28434388.
- Tantawy M, Elzayat MG, Yehia D, et al. Identification of microRNA signature in different pediatric brain tumors. Genet Mol Biol. 2018 Jan–Mar;41(1):27–34. PubMed PMID: 29658967; PubMed Central PMCID: PMCPMC5901491.
- Park MH. The post-translational synthesis of a polyamine-derived amino acid, hypusine, in the eukaryotic translation initiation factor 5A (eIF5A). J Biochem. 2006 Feb;139(2):161–169. PubMed PMID: 16452303; PubMed Central PMCID: PMCPMC2494880.
- Taylor CA, Zheng Q, Liu Z, et al. Role of p38 and JNK MAPK signaling pathways and tumor suppressor p53 on induction of apoptosis in response to Ad-eIF5A1 in A549 lung cancer cells. Mol Cancer. 2013 May 2;12:35. PubMed PMID: 23638878; PubMed Central PMCID: PMCPMC3660295.
- Bussard KM, Mutkus L, Stumpf K, et al. Tumor-associated stromal cells as key contributors to the tumor microenvironment [journal article]. Breast Cancer Res. 2016 August 11;18(1):84.
- Guan XY, Fung JM, Ma NF, et al. Oncogenic role of eIF-5A2 in the development of ovarian cancer. Cancer Res. 2004 Jun 15;64(12):4197–4200. PubMed PMID: 15205331.
- Wei JH, Cao JZ, Zhang D, et al. EIF5A2 predicts outcome in localised invasive bladder cancer and promotes bladder cancer cell aggressiveness in vitro and in vivo. Br J Cancer. 2014 Apr 2;110(7):1767–1777. PubMed PMID: 24504366; PubMed Central PMCID: PMCPMC3974079.
- Bao Y, Lu Y, Wang X, et al. Eukaryotic translation initiation factor 5A2 (eIF5A2) regulates chemoresistance in colorectal cancer through epithelial mesenchymal transition. Cancer Cell Int. 2015;15:109. PubMed PMID: 26581310; PubMed Central PMCID: PMCPMC4650515.
- Eckerdt F, Beauchamp E, Bell J, et al. Regulatory effects of a Mnk2-eIF4E feedback loop during mTORC1 targeting of human medulloblastoma cells. Oncotarget. 2014 Sep 30;5(18):8442–8451. PubMed PMID: 25193863; PubMed Central PMCID: PMCPMC4226695.
- Brummer A, Hausser J. MicroRNA binding sites in the coding region of mRNAs: extending the repertoire of post-transcriptional gene regulation. Bioessays. 2014 Jun;36(6):617–626. PubMed PMID: 24737341.
- Suzuki H, Maruyama R, Yamamoto E, et al. Epigenetic alteration and microRNA dysregulation in cancer. Front Genet. 2013 Dec 3;4:258. PubMed PMID: 24348513; PubMed Central PMCID: PMCPMC3847369.
- Zakraoui O, Marcinkiewicz C, Aloui Z, et al. Lebein, a snake venom disintegrin, suppresses human colon cancer cells proliferation and tumor-induced angiogenesis through cell cycle arrest, apoptosis induction and inhibition of VEGF expression. Mol Carcinog. 2017 Jan;56(1):18–35. PubMed PMID: 26824338.
- Asghar U, Witkiewicz AK, Turner NC, et al. The history and future of targeting cyclin-dependent kinases in cancer therapy. Nat Rev Drug Discov. 2015 Feb;14(2):130–146. PubMed PMID: 25633797; PubMed Central PMCID: PMCPMC4480421.
- Gonzalez-Vera JA, Bouzada D, Bouclier C, et al. Lanthanide-based peptide biosensor to monitor CDK4/cyclin D kinase activity. Chem Commun (Camb). 2017 Jun 1;53(45):6109–6112. PubMed PMID: 28530267.
- Whittaker SR, Barlow C, Martin MP, et al. Molecular profiling and combinatorial activity of CCT068127: a potent CDK2 and CDK9 inhibitor. Mol Oncol. 2018 Mar;12(3):287–304. PubMed PMID: 29063678; PubMed Central PMCID: PMCPMC5830651.
- Opferman JT, Kothari A. Anti-apoptotic BCL-2 family members in development. Cell Death Differ. 2018 Jan;25(1):37–45. PubMed PMID: 29099482; PubMed Central PMCID: PMCPMC5729530.
- Siddiqui WA, Ahad A, Ahsan H. The mystery of BCL2 family: Bcl-2 proteins and apoptosis: an update. Arch Toxicol. 2015 Mar;89(3):289–317. PubMed PMID: 25618543.
- Reed JC, Stein C, Subasinghe C, et al. Antisense-mediated inhibition of BCL2 protooncogene expression and leukemic cell growth and survival: comparisons of phosphodiester and phosphorothioate oligodeoxynucleotides. Cancer Res. 1990 Oct 15;50(20):6565–6570. PubMed PMID: 2208117.
- Sun SY, Yue P, Zhou JY, et al. Overexpression of BCL2 blocks TNF-related apoptosis-inducing ligand (TRAIL)-induced apoptosis in human lung cancer cells. Biochem Biophys Res Commun. 2001 Jan 26;280(3):788–797. PubMed PMID: 11162590.
- Lin YC, Lin JF, Tsai TF, et al. Tumor suppressor miRNA-204-5p promotes apoptosis by targeting BCL2 in prostate cancer cells. Asian J Surg. 2017 Sep;40(5):396–406. PubMed PMID: 27519795.
- Zhu W, Cai MY, Tong ZT, et al. Overexpression of EIF5A2 promotes colorectal carcinoma cell aggressiveness by upregulating MTA1 through C-myc to induce epithelial-mesenchymaltransition. Gut. 2012 Apr;61(4):562–575. PubMed PMID: 21813470.
- Meng QB, Kang WM, Yu JC, et al. Overexpression of eukaryotic translation initiation factor 5A2 (EIF5A2) correlates with cell aggressiveness and poor survival in gastric cancer. PloS one. 2015;10(3):e0119229. PubMed PMID: 25793713; PubMed Central PMCID: PMC4368542.
