ABSTRACT
Rhodococcus sp. 2N was found as a 1,3-propanediols-oxidizing strain from soil samples through enrichment culture using 2,2-diethyl-1,3-propanediol (DEPD) as the sole carbon source. The culture condition of the strain 2N was optimized, and the highest activity was observed when 0.3% (w/v) DEPD was added in the culture medium as an inducer. Chiral HPLC analysis of the hydroxyalkanoic acid converted from 2-ethyl-2-methyl-1,3-propanediol (EMPD) revealed that the strain 2N catalyzed the (R)-selective oxidation of EMPD. The reaction products and intermediates from DEPD and EMPD were identified by nuclear magnetic resonance analyses, and the results suggested that only one hydroxymethyl group of the propanediols was converted to carboxy group via two oxidation steps. Under optimized conditions and after a 72-h reaction time, the strain 2N produced 28 mM (4.1 g/L) of 2-(hydroxymethyl)-2-methylbutanoic acid from EMPD with a molar conversion yield of 47% and 65% ee (R).
Graphical Abstract
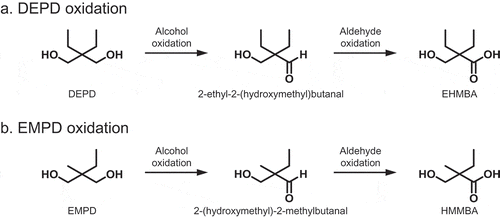
Enzymatic pathways for the oxidation of DEPD or EMPD to the corresponding hydroxyalkanoic acids via aldehydes by whole cells of Rhodococcus sp. 2N.
Optically active hydroxyalkanoic acids are beneficial as pharmaceutical intermediates, chiral synthons, and monomers for functional polymers, and various studies have been performed to develop an efficient synthesis method of valuable hydroxyalkanoic acids [Citation1–Citation3]. Hydroxyalkanoic acids are enzymatically synthesized through the following two unique reactions: 1) hydroxylation of the terminal methyl group of alkanoic acids and 2) oxidation of the unilateral hydroxymethyl group of diols. For example, the (R)- and (S)-enantiomers of 3-hydroxy-2-methylpropanoic acid can be synthesized from 2-methylpropanoic acid by microbial asymmetric hydroxylation [Citation4–Citation6]. The microbial asymmetric oxidation of 2-methyl-1,3-propanediol by acetic acid bacteria Gluconobacter and Acetobacter strains produce (R)-3-hydroxy-2-methylpropanoic acid with high optical purity [Citation7–Citation9]. (R)- and (S)-2-(Hydroxymethyl)hexanoic acid, which comprise useful constituents of an anti-bacterial agent for multidrug-resistant Staphylococcus aureus [Citation10,Citation11], are produced by Pseudomonas putida IFO 3738 and A. pasteurianus IAM 12073, respectively [Citation12]. However, to our knowledge, the production of hydroxyalkanoic acids from 2,2-disubstituted-1,3-propanediols has not yet been studied, and the microbial enzymes accepting various propanediols as substrates have not been reported.
Various hydroxybutanoic acids are useful as configurational molecular glues [Citation13–Citation15]. 2-(Hydroxymethyl)-2-methylbutanoic acid (HMMBA) can be produced by 2-ethyl-2-methyl-1,3-propanediol (EMPD) oxidation; it has significant potential as raw material for the synthesis of a novel functional polymer. In this study, we report the screening of microorganisms oxidizing 2,2-diethyl-1,3-propanediols (DEPD) and the synthesis of hydroxyalkanoic acids, especially chiral HMMBA, using whole cells of a DEPD-oxidizing strain ().
Materials and methods
Materials
The diol compounds, DEPD and EMPD, were purchased from Tokyo Chemical Industry Co., Ltd. (Japan). Products from the following suppliers were used: Polypepton (Nippon Pharmaceutical Co., Ltd., Japan), meat extract (Kyokuto Pharmaceutical Industrial Co., Ltd., Japan), and yeast extract (Oriental Yeast Co., Ltd., Japan). All other chemicals were certified to be of reagent grade.
Microorganisms and culture conditions
The soil samples were added to 80 mL of enrichment culture medium comprising 0.5% (w/v) DEPD, 0.3% (w/v) NH4Cl, 0.2% (w/v) K2HPO4, 0.1% (w/v) NaCl, 0.01% (w/v) MgSO4·7H2O, 0.001% (w/v) FeSO4·7H2O, 1% (v/v) vitamin mixture, and 0.1% (v/v) metal mixture. Microorganisms were isolated on the agar plate of enrichment culture medium. The vitamin mixture consisted of 0.01% (w/v) biotin, 0.01% (w/v) inositol, 0.002% (w/v) calcium pantothenate, 0.002% (w/v) nicotinic acid, 0.002% (w/v) pyridoxine·HCl, 0.001% (w/v) p-aminobenzoic acid, 0.001% (w/v) riboflavin, and 1.1 μM folic acid. The metal mixture consisted of 0.04% (w/v) CaCl2·2H2O, 0.03% (w/v) H3BO3, 0.004% (w/v) CuSO4·5H2O, 0.01% (w/v) KI, 0.02% (w/v) FeSO4·7H2O, 0.04% (w/v) MnSO4·7H2O, and 0.02% (w/v) NaMoO4·2H2O in 1% (v/v) conc. HCl. The soil isolates were cultivated in 50 mL of a nutrient medium comprising 0.5% (w/v) Polypepton, 0.5% (w/v) meat extract, 0.2% (w/v) NaCl, and 0.05% (v/v) yeast extract in 500 mL shake flasks. The isolates were cultured at 28 °C for 2 days with reciprocal shaking (120 strokes/min).
The genome of the soil isolate strain 2N was extracted and purified, and the 16S rRNA gene was amplified and cloned into the pT7Blue vector. The 16S rRNA gene sequencing analysis was performed with the primers 27f (5ʹ-AGAGTTTGATCCTGGCTCAG-3ʹ) and 1525r (5ʹ-AAAGGAGGTGATCCAGCC-3ʹ), the GenomeLab DTCS Quick Start Kit (Sciex, USA), and a Beckman CEQ8000 DNA sequencer (Beckman Coulter, UK).
Propanediol oxidation reaction using whole cells
The oxidation of propanediols was carried out in 4 mL of reaction mixtures comprising 10 mM propanediols, 100 mM potassium phosphate (KPi) buffer (pH 8.0), and whole cells of the soil isolate Rhodococcus sp. 2N (11.9 mg dry cell weight) in 50 mL sample tube (Maruemu Corp., Japan) at 30 °C with reciprocal shaking (120 strokes/min). The strain 2N was precultivated at 28 °C for a day, then the whole broth was inoculated into 50 mL of the nutrient medium described above. The whole cells of the strain 2N were harvested by centrifugation, washed with 0.85% (w/v) NaCl twice, and resuspended with 0.85% (w/v) NaCl.
The effect of pH condition was investigated using 100 mM of the following buffers: citrate-sodium citrate (pH 5.0–6.0), acetate-sodium acetate (pH 5.0–5.5), KPi (pH 6.0–8.0), Tris-HCl (pH 7.5–9.0), and glycine-NaOH (pH 9.0–10.0). The effect of temperature was investigated at 20–40 °C.
Analyses and identification of the reaction products
The reaction products were analyzed with thin-layer chromatography (TLC) using 2,6-dichlorophenolindophenol for carboxy group detection. The products and intermediates were also analyzed by high-performance liquid chromatography (HPLC) with a YMC-Triart C18 column (4.6 × 150 mm; YMC Co., Ltd.) coupled to an SPD-10AVP UV-Vis detector (Shimadzu, Japan). The eluent, 50 mM NaH2PO4/H3PO4 (pH 2.8)/acetonitrile (4:1 v/v), was applied at a flow rate of 1 mL/min and the eluate was monitored at 210 nm. The reaction products were extracted with diethyl ether after adjusting the pH of the reaction mixture to pH 3 with 2 M HCl. The crude products were purified by silica gel column chromatography (30 × 110 mm, Wakogel C-300; FUJIFILM Wako Pure Chemical Co., Japan), and the following eluents (v/v) were run in sequence: n-hexane only (200 mL), n-hexane/ethyl acetate = 6:1 (350 mL), n-hexane/ethyl acetate = 4:1 (300 mL), and n-hexane/ethyl acetate = 2:1 (600 mL).
The chemical shifts of 2-ethyl-2-(hydroxymethyl)butanoic acid (EHMBA) were determined: 1H-NMR (600 MHz, D2O); δ 0.59 (3H × 2, t, J = 7.2 Hz, -CH2-CH3), 1.29 (2H × 2, q, J = 7.7 Hz, -CH2-CH3), and 3.47 (2H, s, -CH2-OH), and 13C-NMR (150 MHz, D2O); δ 8.23, 26.24, 52.56, 61.44, and 184.34. The chemical shifts of HMMBA were determined: 1H-NMR (600 MHz, D2O); δ 0.64 (3H, t, J = 7.6 Hz, -CH2-CH3), 0.89 (3H, s, -CH3), 1.29 (2H, dq, J = 7.6 Hz, 7.6 Hz, -CH2-CH3), 3.33 (1H, d, J = 11 Hz, -CH2-OH), and 3.46 (1H, d, J = 11 Hz, -CH2-OH), and 13C-NMR (150 MHz, D2O); δ 8.43, 19.50, 28.40, 49.08, 67.85, and 185.10.
The intermediates were extracted with diethyl ether and derivatized with 15 mM 2,4-dinitrophenylhydrazine to form hydrazone derivatives [Citation16]. The hydrazone derivatives were fractionated and purified on PLC Silica gel 60 F254, 2 mm (Merck, Germany) with ethyl acetate/n-hexane = 4:1. The identification was carried out with 1H-NMR and 13C-NMR.
Optical purity of HMMBA
The HMMBA produced from EMPD was derivatized with phenacyl bromide according to the manufacturer’s protocol (TCI, Japan). The optical purity of the obtained derivative was evaluated with chiral HPLC analysis on a ChiralPak AD-H column (4.6 × 150 mm; Daicel Corp., Japan) coupled to an SPD-10AVP UV-Vis detector (Shimadzu). The eluent, n-hexane/ethanol (7:3 v/v), was applied at a flow rate of 1 mL/min, and the eluate was monitored at 254 nm. (S)- and (R)-HMMBAs were eluted separately and analyzed in comparison with the elution of authentic compounds. The retention times of (S)- and (R)-HMMBAs were 11.2 and 13.6 min, respectively.
Results
Isolation and selection of DEPD-oxidizing microorganisms
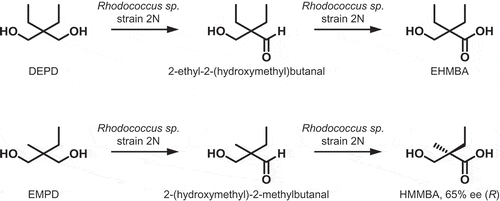
To establish the efficient synthesis of optically active hydroxyalkanoic acids using microbial enzymes, we isolated 30 strains of DEPD-consuming microorganisms through enrichment culture with medium comprising DEPD as the sole carbon source. Three strains of these DEPD-consuming microorganisms were selected for further analysis based on the spot intensity of carboxylic acids on TLC analysis. After a 24-h incubation of the whole cells with DEPD, the formation of monocarboxylic acid and dicarboxylic acid was evaluated by HPLC analysis. Among the strains, only one, named as 2N, was able to synthesize monocarboxylic acid without forming dicarboxylic acid, indicating the oxidization of the unilateral hydroxymethyl group of DEPD. The NMR analysis of the isolated monocarboxylic acid [retention time (r.t.) = 5.57 on HPLC analysis; Rf = 0.48 on TLC using ethyl acetate] revealed that the monocarboxylic acid was EHMBA. The strain 2N exhibited colonial morphology of smooth surface, moist texture, and pink color. The 16S rRNA gene (1,511 bp, accession no. LC434455) sequencing analysis of strain 2N showed 100% identity to Rhodococcus sp. JCM 28273 (accession no. LC133620) and Rhodococcus sp. S10 (accession no. MG551531), and the strain 2N was identified as Rhodococcus sp. 2N.
The whole cells of Rhodococcus sp. 2N also catalyzed the oxidation of EMPD. The product was isolated and analyzed by NMR using the same method applied to DEPD. The product (r.t. = 3.70 on HPLC analysis; Rf = 0.75 on TLC using ethyl acetate/methanol = 4:1) converted from EMPD was identified as HMMBA. The product HMMBA was derivatized with phenacyl bromide and analyzed with chiral HPLC. The results showed that the whole-cell reaction exhibited (R)-selectivity (65% ee) in the oxidation of EMPD.
Identification of the intermediates in the oxidation reaction
In the early stage of the oxidizing reaction of either DEPD or EMPD, an unknown product was identified on HPLC analysis. The unknown products from DEPD and EMPD disappeared in accordance with the formation of the corresponding hydroxyalkanoic acids. Thus, it was suggested that the unknown products were the aldehydes formed during the hydroxybutanoic acids formation. The unknown products were extracted with diethyl ether and derivatized with 2,4-dinitrophenylhydrazine to form 2,4-dinitrophenylhydrazone derivatives. The derivatives were obtained as yellow solids, then washed with pure water to thoroughly remove the HCl, and analyzed with NMR. The unknown product from DEPD was identified as 2-ethyl-2-(hydroxymethyl)butanal and that from EMPD was identified as 2-(hydroxymethyl)-2-methylbutanal, as shown in .
The 2,4-dinitrophenylhydrazone derivative of 2-ethyl-2-(hydroxymethyl)butanal (r.t. = 9.32 on HPLC analysis; Rf = 0.47 on TLC using ethyl acetate/n-hexane = 4:5): 1H-NMR (600 MHz, CDCl3); δ 0.90 (3H × 2, t, J = 7.6 Hz, -CH2-CH3), 1.63 (2H × 2, q, J = 7.6 Hz, -CH2-CH3), 1.77 (1H, t, J = 5.9 Hz, -OH), 3.74 (2H, d, J = 3.4 Hz, -CH2-OH), 7.43 (1H, s, -C(= N-)-H), 7.85 (1H, d, J = 9.6 Hz, -NH-C-CH-CH-), 8.31 (1H, dd, J = 2.8, 9.6 Hz, -NH-C-CH-CH-C(NO2)-), 9.12 (1H, d, J = 2.8 Hz, -NH-C-C(NO2)-CH-C(NO2)-), and 11.04 (1H, s), and 13C-NMR (150 MHz, CDCl3); δ 7.85, 25.07, 46.58, 65.03, 116.29, 123.55, 128.96, 130.10, 137.94, 144.96, and 157.28. The 2,4-dinitrophenylhydrazone derivative of 2-(hydroxymethyl)-2-methylbutanal (r.t. = 5.28 on HPLC analysis; Rf = 0.41 on TLC using ethyl acetate/n-hexane = 1:1.25): 1H-NMR (600 MHz, CDCl3); δ 0.95 (3H, t, J = 7.6 Hz, -CH2-CH3), 1.19 (3H, s), 1.59–1.67 (2H, m, -CH2-CH3), 1.89 (1H, s, -OH), 3.65 (1H, d, J = 11.0 Hz, -CH2-OH), 3.76 (1H, d, J = 11.0, -CH2-OH), 7.48 (1H, s, -C(= N-)-H), 7.88 (1H, d, J = 9.7 Hz, -NH-C-CH-CH), 8.32 (1H, dd, J = 2.8, 9.6 Hz, -NH-C-CH-CH-C(NO2)-), 9.13 (1H, d, J = 2.8 Hz, -NH-C-C(NO2)-CH-C(NO2)-), and 11.05 (1H, s), and 13C-NMR (150 MHz, CDCl3); δ 8.18, 19.10, 28.40, 43.90, 68.14, 116.32, 123.51, 128.93, 130.06, 137.91, 144.99, and 157.33.
Enzyme induction, optimization of the reaction conditions, and production of hydroxyalkanoic acids
The alcohol-oxidizing activity of the strain 2N was inducibly expressed by the addition of various propanediols. No activity was detected without propanediols in the culture medium (). The addition of 0.3% (w/v) DEPD induced the highest level of alcohol-oxidizing activity, whereas the addition of 2-amino-1,3-propanediols induced no oxidizing activity.
Table 1. Induction of oxidation activity by propanediol.
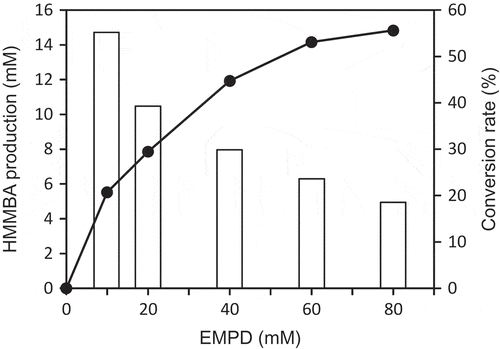
The 2N cells exhibited the highest oxidation activity when cultured for 60 h at 28 °C in the nutrient medium comprising 0.3% (w/v) DEPD, pH 7.0. The highest activity was observed when the whole-cell reaction was performed at 30 °C in 100 mM KPi buffer, pH 8.0, with reciprocal shaking at 120 strokes/min. In the 100 mM KPi (pH 7.0), the oxidation activity was about 70% of that in 100 mM KPi (pH 8.0). The oxidizing activity of the strain 2N cells was kept well in the presence of less than 60 mM EMPD (), indicating that excess addition of EMPD inhibits the oxidation activity.
Figure 2. Effect of EMPD concentration on HMMBA production (solid circle) and conversion rate (open bar).
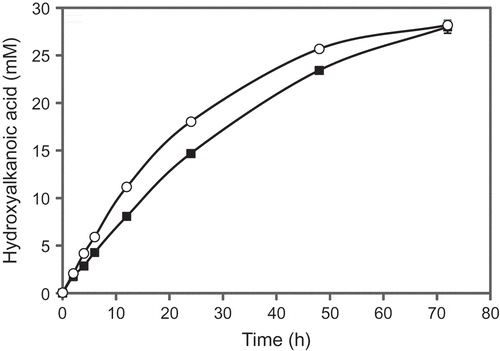
The conversion of DEPD and EMPD to EHMBA and HMMBA, respectively, was performed under the optimal conditions to achieve efficient production of the hydroxyalkanoic acids (). The productivity of EHMBA reached 28 mM (4.1 g/L) with a molar conversion yield of 47% and 28 mM (4.1 g/L) HMMBA was formed from 60 mM EMPD with a molar conversion yield of 47% and 65% ee (R). The final pH of the reaction supernatant was around pH 7.0.
Substrate specificity of the oxidizing enzyme(s) in Rhodococcus sp. 2N
Substrate specificity of the oxidizing enzyme(s) was investigated using the whole cells of the strain 2N and 10 mM various 1,3-propanediols (). The strain 2N cells accepted 2-methyl-1,3-propanediol, 2,2-dimethyl-1,3-propanediol, and 2-methyl-2-propyl-1,3-propanediol besides DEPD and EMPD. The activity was 1.66-fold higher for 2-methyl-1,3-propanediol than for DEPD. The strain 2N cells did not accept 1,3-propanediols comprising a butyl group, hydroxymethyl group, methylene group, or amino group in the 2-position, such as 1,3-propanediol, 2-butyl-1,3-propanediol, glycerol, 2-phenyl-1,3-propanediol, 2-methylene-1,3-propanediol, 2-butyl-2-ethyl-1,3-propanediol, 2-hydroxymethyl-2-methyl-1,3-propanediol, 2-amino-1,3-propanediol, 2-amino-2-methyl-1,3-propanediol, and 2-amino-2-hydroxymethyl-1,3-propanediol.
Table 2. Substrate specificity of the oxidizing enzyme(s) in Rhodococcus sp. 2N cells.
Discussion
We isolated and screened microorganisms through enrichment culture comprising DEPD as a sole carbon source. We identified one strain, Rhodococcus sp. 2N, capable of catalyzing the asymmetric oxidation of EMPD to (R)-HMMBA. Several diol dehydrogenases from Rhodococcus sp. are reported as NAD+/NADH-dependent enzymes [Citation17,Citation18]. With respect to the Rhodococcus sp. 2N, the cell-free extract exhibits no oxidation activity upon the addition of coenzymes NAD+, NADP+, NADH, and NADPH (data not shown). For the characterization of coenzyme dependence, enzyme(s) identification and production of recombinant enzyme(s) are required to clarify the coenzyme requirement. The oxidation activity of the strain 2N cells was highly induced on addition of 0.3% (w/v) DEPD to the culture medium. Conversely, the addition of >0.4% (w/v) DEPD decreased the oxidation activity. The oxidation activity was less induced by the propanediols with lower steric hindrance than DEPD, such as 2-methyl-1,3-propanediol, 2,2-dimethyl-1,3-propanediol, and EMPD. The aminopropanediols barely induced the activity.
During the conversion of DEPD and EMPD into their corresponding hydroxymethyl carboxylic acids using the strain 2N cells, the formation of the aldehyde intermediates 2-ethyl-2-(hydroxymethyl)butanal and 2-(hydroxymethyl)-2-methylbutanal was observed, respectively. In previous studies [Citation7–Citation12], no aldehyde intermediate was detected. Presumably, the dehydrogenase(s) involving in the asymmetric oxidation of the present strain 2N have characteristics different from the membrane-bound dehydrogenases of acetic acid bacteria [Citation19]. In the oxidation reaction of their membrane-bound dehydrogenases, no aldehyde intermediate was detected because the alcohol substrate can be immediately transferred to following enzyme aldehyde dehydrogenase. The dehydrogenase(s) were soluble in cell-free extract, so the aldehyde intermediate might be discharged from the conversion flow. The chemical synthesis of the abovementioned aldehydes has been reported [Citation20]; however, their enzymatic or microbial synthesis remains to be reported. By inhibiting the aldehyde oxidation involved in the subsequent oxidation reaction of Rhodococcus sp. 2N by gene modification or with inhibitor for the dehydrogenase(s), the aldehydes might be exclusively produced.
In the conversion of EMPD, the oxidation activity of the strain 2N cells decreased with over 60 mM EMPD. During the oxidation of DEPD and EMPD (), both DEPD and EMPD were converted with a molar conversion yield of 47%. The final pH of the reaction was around pH 7.0, indicating that the dehydrogenase(s) in the strain 2N was not deactivated by the pH decrease because the enzyme(s) can work at pH 7.0 as described in the Results section. These results suggest that the oxidation activities of the strain 2N cells are inhibited by the produced EHMBA and HMMBA.
In the present study, we found Rhodococcus sp. 2N that synthesizes EHMBA and (R)-HMMBA from DEPD and EMPD, respectively. To isolate this strain, we used DEPD as a sole carbon source for enrichment culture. Enrichment culture using EMPD instead of DEPD would lead to acquire a much higher number of microorganisms oxidizing various 1,3-propanediols because EMPD has less steric hindrance than DEPD. To achieve the production of S-form of HMMBA, a promising molecular glue of polymers for development of novel functional materials or a building block for chemical synthesis, we need to further isolate microorganisms capable of catalyzing the asymmetric oxidation of 1,3-propanediols. Furthermore, to establish a more efficient asymmetric oxidation system, we would need to identify the genes encoding the alcohol dehydrogenase(s) and aldehyde dehydrogenase(s) oxidizing the 1,3-propanediols in the strain 2N.
Author contributions
K. Mitsukura and H. Kikukawa designed the research. H. Kikukawa, R. Koyasu, and K. Mitsukura performed the experiments. H. Kikukawa wrote the paper. T. Yoshida supervised all the experiments and data analyses.
Acknowledgments
We would like to thank Enago (www.enago.jp) for English language editing.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- De Vitis V, Dall’Oglio F, Pinto A, et al. Chemoenzymatic synthesis in flow reactors: a rapid and convenient preparation of captopril. ChemistryOpen. 2017;6(5):668–673.
- Ren Q, Grubelnik A, Hoerler M, et al. Bacterial poly(hydroxyalkanoates) as a source of chiral hydroxyalkanoic acids. Biomacromolecules. 2005;6(4):2290–2298.
- Singh AK, Mallick N. Advances in cyanobacterial polyhydroxyalkanoates production. FEMS Microbiol Lett. 2017;364(20):fnx189.
- Goodhue CT, Schaeffer JR. Preparation of L(+)-β-hydroxyisobutyric acid by bacterial oxidation of isobutyric acid. Biotechnol Bioeng. 1971;13:203–214.
- Hasegawa J, Hamaguchi S, Ogura M, et al. Studies on β-hydroxylation acids II. Production of β-hydroxyisobutyric acids from aliphatic carboxylic acids by microorganisms. J Ferment Technol. 1981;59:257–262.
- Hasegawa J, Ogura M, Hamaguchi S, et al. Studies on β-hydroxylation acids I. Stereoselective conversion of isobutyric acid to β-hydroxyisobutyric acid by microorganisms. J Ferment Technol. 1981;59:203–208.
- Leon R, Prazeres DM, Fernandes P, et al. A multiphasic hollow fiber reactor for the whole-cell bioconversion of 2-methyl-1,3-propanediol to (R)-β-hydroxyisobutyric acid. Biotechnol Prog. 2001;17(3):468–473.
- Molinari F, Gondolfi R, Villa R, et al. Enantioselective oxidation of prochiral 2-methyl-1,3-propanediol by Acetobacter pasteurianus. Tetrahedron-Asymmetry. 2003;14:2041–2043.
- Ohta H, Tetsukawa H, Noto N. Enantiotopically selective oxidation of α,ω-diols with the enzyme systems of microorganisms. J Org Chem. 1982;47:2400–2404.
- Fujita M, Sakamoto M, Horiuchi N, et al. inventors; Dainippon Pharmaceutical Co., Ltd., assignee. Proline derivatives. PCT Int Appl. WO2004018453A1. 2004.
- Kapa PK, Jiang XL, Loeser EM, et al., inventors; Novartis Ag and Novartis Pharma Gmbh, asignee. Process for preparing intermediates. PCT Int Appl. WO2004026824A1. 2004.
- Mitsukura K, Uno T, Yoshida T, et al. Microbial asymmetric oxidation of 2-butyl-1,3-propanediol. Appl Microbiol Biotechnol. 2007;76(1):61–65.
- Tsuji H, Noda S, Kimura T, et al. Configurational molecular glue: one optically active polymer attracts two oppositely configured optically active polymers. Sci Rep. 2017;7:45170.
- Tsuji H, Sobue T. Cocrystallization of monomer units in lactic acid-based biodegradable copolymers, poly(l-lactic acid-co-l-2-hydroxybutanoic acid)s. Polymer. 2015;72:202–211.
- Tsuji H, Sobue T. Stereocomplex crystallization and homo-crystallization of enantiomeric substituted poly(lactic acid)s, poly(2-hydroxy-3-methylbutanoic acid)s. Polymer. 2015;69:186–192.
- Yamada M, Okada Y, Yoshida T, et al. Purification, characterization and gene cloning of isoeugenol-degrading enzyme from Pseudomonas putida IE27. Arch Microbiol. 2007;187(6):511–517.
- Baer K, Krausser M, Burda E, et al. Sequential and modular synthesis of chiral 1,3-diols with two stereogenic centers: access to all four stereoisomers by combination of organo- and biocatalysis. Angew Chem Int Ed Engl. 2009;48(49):9355–9358.
- van der Werf MJ, Swarts HJ, de Bont JA. Rhodococcus erythropolis DCL14 contains a novel degradation pathway for limonene. Appl Environ Microbiol. 1999;65(5):2092–2102.
- Yakushi T, Matsushita K. Alcohol dehydrogenase of acetic acid bacteria: structure, mode of action, and applications in biotechnology. Appl Microbiol Biotechnol. 2010;86(5):1257–1265.
- Culp RD, Beavers WA, inventors; Eastman Chemical Company, asignee. Preparation of an aldol using a base-modified clay catalyst. PCT Int Appl. WO2000000456A1. 2000.