ABSTRACT
Sake yeast was first isolated as a single yeast strain, Saccharomyces sake, during the Meiji era. Yeast strains suitable for sake fermentation were subsequently isolated from sake brewers and distributed as Kyokai yeast strains. Sake yeast strains that produce characteristic flavors have been bred in response to various market demands and individual preferences. Interestingly, both genetic and morphological studies have indicated that sake yeast used during the Meiji era differs from new sake yeasts derived from Kyokai Strain No. 7 lineage. Here, we discuss the history of sake yeast breeding, from the discovery of sake yeast to the present day, to highlight the achievements of great Japanese scientists and engineers.
Graphical Abstract
History of sake yeast started with the isolation of Saccharomyces sake in the Meiji era.
Sake yeast (Saccharomyces sake) was first isolated in 1895, during the Meiji era (1868–1912). During this same year, Japan won the Sino-Japanese War and concluded the Nissin Peace Treaty (Shimonoseki Convention); Ichiyō Higuchi published her famous novel “Takekurabe”; and a distinguished two-way baseball star player, Babe Ruth, was born. Since then, sake yeast has become indispensable for sake brewing, largely through cooperation between the public and private sectors, to select and breed strains with favorable characteristics. Several types of premium-grade sake are now produced and distributed in various regions throughout Japan. A national sake appraisal meeting is held annually to further improve the quality of sake.
Given that drinking sake is an established Japanese tradition, the development of sake yeasts used for sake brewing is a matter of cultural importance in Japan. In this article, we discuss the history of sake yeasts, from their discovery to the present. We also evaluate their development based on phenotypic and genotypic analyses.
Discovery of Saccharomyces sake
Until the Edo period, the raw materials used to make sake mash included only steamed rice, water, and koji mold. Brewed yeast naturally grows in the sake mash, producing alcohol. However, no brewers in the Edo period identified brewed yeast in the sake mash. Saccharomyces cerevisiae itself was first isolated by Franz Julius Ferdinand Meyen in 1837 and was later given its scientific name by Emil Christian Hansen in 1883 [Citation1,Citation2]. It later became well-known that beer and wine are fermented by budding yeast [Citation3]. However, until the early Meiji era, it was not known that budding yeast is also involved in the alcoholic fermentation of sake. Other types of alcohol are fermented by different types of fungus. For example, in Shaoxing wine (known as Yellow wine in China) Rhizopus spp. is responsible for alcohol fermentation. A German paper also reported that alcoholic fermentation of sake was achieved by Aspergillus spp [Citation4]. The isolation of sake yeast in Japan disproved this theory.
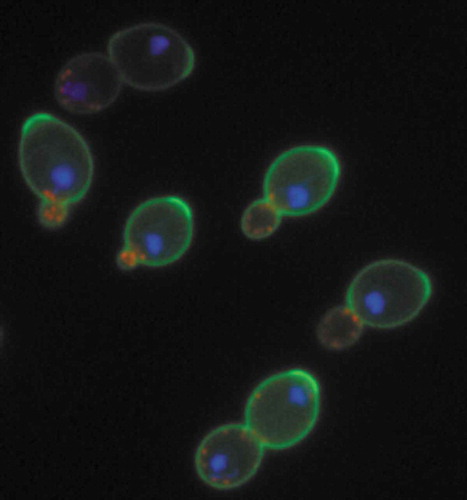
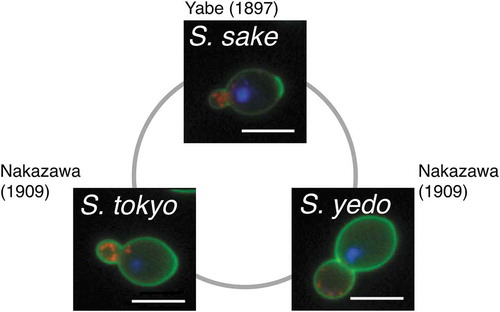
Sake yeast was observed in a sake brewery in 1881 by Robert William Atkinson of the University of Tokyo [Citation5]. Kikuji Yabe (First Primer of Brewery Research Institute) and Yoshinao Kozai (Professor of the Tokyo Imperial University Agricultural University), famous for demonstrating copper contamination in the Ashio Mineral Poison Incident, reported in 1895 that they had isolated yeast (Saccharomyces sake YABE et KOZAI) from rice koji [Citation6–Citation8]. Soon after, Ryoji Nakazawa of the National Research Institute of Brewing (NRIB) analyzed two kinds of sake yeast, S. tokyo and S. yedo, at the Munich Brewing Institute in Germany () [Citation9]. This represented the beginning of research on sake yeast by Japanese scientists.
Figure 1. Early research on sake yeast. The cell wall (green), actin (red) and nuclear DNA (blue) of S. sake, S. tokyo and S. yedo were stained and photographed. Bars indicate 5 µm.
However, there are several reasons to be skeptical about the veracity of this historical anecdote. For example, it is not clear whether rice koji is a good source of sake yeast [Citation7]. If rice koji is left exposed to the external environment, it is likely to be colonized by naturally occurring yeasts. This principle is similar to the collection of natural yeasts for bread-making by exposing sugar-pickled fruit to the air [Citation10]. Isolating sake yeast from fermenting sake mash would have been a more reliable approach. Unfortunately, the original paper does not report whether S. sake was used to make sake [Citation7,Citation11]. S. tokyo and S. yedo were originally sent to the Munich Brewing Institute from Japan several years ago by a Japanese scientist named Ichikawa, but the source of these isolates is unknown [Citation9,Citation11,Citation12]. It is also intriguing that two strains, S. tokyo and S. yedo, were present in the original stock. However, great researchers do not make fundamental errors of fact, even if the details of the surrounding narrative are somewhat ambiguous. At the time of their discovery, S. sake, S. tokyo and S. yedo were classified as different species derived from the budding yeast S. cerevisiae. After the classification and rearrangement of extensive yeast species, all were later re-classified as a single species, S. cerevisiae [Citation13–Citation16]. However, subsequent genomic analyses of many different strains of yeast confirmed the existence of intra-specific variation in S. cerevisiae. Sake yeast is an intraspecific strain that differs significantly from other budding yeasts, such as beer yeast, wine yeast, baker’s yeast and shochu yeast [Citation17–Citation21].
Kyokai Strains no. 1 to no. 5
When sake is fermented using naturally occurring yeasts, its taste and aroma are unstable and are not always satisfactory. Therefore, Professor Teizo Takahashi, who took over Yoshinao Kozai’s Laboratory, devised a plan to cultivate pure sake yeast obtained from high-quality sake breweries. Takahashi consulted with officers in the Tax Supervision Bureau throughout Japan, collected the sake mash from which reputable sakes were made, and isolated more than 60 strains of yeast [Citation22]. Takahashi collaborated with many researchers and analyzed these strains with respect to their cell shape, endospore formation, film formation, fermentation ability, amount of acid and alcohol production, amount of amino acid consumption, and other variables. The resulting body of experimental results was published in 1914 [Citation22]. The researchers noticed that the addition of monocultured yeast isolated from Sakura Masammune in Nada, Hyogo significantly improved the flavor of sake. They started to distribute this “pure cultured yeast” (Kyokai Strain No. 1), and this was the birth of the Kyokai yeast [Citation11,Citation23].
High-quality sake yeasts were subsequently obtained from Gekkeikan in Fushimi, Kyoto (Kyokai Strain No. 2) and from sake breweries in Hiroshima prefecture (Kyokai Strains No. 3–5), and were distributed by the Brewing Society of Japan (BSJ). Contemporary national policy has focused on increasing wealth and military power, where one strategy focused on increasing revenue from liquor taxes by producing higher-quality sake. The distribution of pure cultured yeast strains by the BSJ undoubtedly improved the quality of sake, consequently boosting its sales. Interestingly, a total of eight strains from S. sake, S. tokyo and S. yedo, as well as Kyokai Strains No. 1–5 mentioned above, exhibited similar characteristics [Citation24]. Sake brewing yeast, present in Japan from the Meiji era to the Taisho era, likely descended from a comparatively recent common ancestor. A recent study on the genetic lineage of S. cerevisiae species confirmed this theory [Citation25].
The birth of Kyokai Strain no. 6
Despite all of their successes, the BSJ ceased distribution of Kyokai Strains No. 1–5 by 1940. The turning point was in 1935, when the BSJ started distributing a new strain, Kyokai Strain No. 6 (). Kyokai Strain No. 6 possessed favorable characteristics that previous strains did not have, such as high fermentation ability at low temperatures. It was the only strain that was distributed during the Pacific War, from 1941 to 1945. This strain has been distributed for more than 80 years, which represents a long period of continuous use. The closely related Kyokai Strains No. 7, No. 9 and No. 10 began to be distributed in 1946, 1968 and 1977, respectively () [Citation20,Citation24,Citation26].
Figure 2. Tree diagram of sake yeast strains. Tree diagram of sake yeast strains. Rectangles and ovals indicate sake yeast strains analyzed or not analyzed in the study, respectively. Strains in the gray box (new sake yeast, also referred known as Kyokai Strain No. 7, K7 lineage) constitute the group of sake yeasts that putatively share a common ancestral origin. Among the new sake yeast strains, the four strains highlighted in the box (K6, K7, K9 and K10) are the strains used as the original breeding strains. Arrows between strains indicate breeding history (e.g. K601 is isolated from K6); if isolation was conducted by the crossing of two parental strains and/or mutagen treatment (dashed line), both are indicated beside them. The diagram was constructed based on the Japanese literature [Citation20,Citation47].
![Figure 2. Tree diagram of sake yeast strains. Tree diagram of sake yeast strains. Rectangles and ovals indicate sake yeast strains analyzed or not analyzed in the study, respectively. Strains in the gray box (new sake yeast, also referred known as Kyokai Strain No. 7, K7 lineage) constitute the group of sake yeasts that putatively share a common ancestral origin. Among the new sake yeast strains, the four strains highlighted in the box (K6, K7, K9 and K10) are the strains used as the original breeding strains. Arrows between strains indicate breeding history (e.g. K601 is isolated from K6); if isolation was conducted by the crossing of two parental strains and/or mutagen treatment (dashed line), both are indicated beside them. The diagram was constructed based on the Japanese literature [Citation20,Citation47].](/cms/asset/43d44c68-88bf-428b-a3f7-82eb008dee69/tbbb_a_1564620_f0002_c.jpg)
Several highly accomplished engineers were involved in the birth of Kyokai Strain No. 6. This sake yeast was isolated in Akita in the Tohoku area, and one key person involved in this was Masatsune Hanaoka, who is known as the father of sake brewing in Akita. Hanaoka first worked in the Sendai Tax Supervision Bureau, but later took office as the first manager of the Akita Research Institute of Brewing. He developed numerous manufacturing technologies, such as a method of improving polished rice to enhance sake flavor, as well as improved techniques for making sake starters and Akita-style koji tray [Citation27–Citation30]. He also investigated low-temperature, long-term fermentation methods suitable for the climate of Akita, helping to prevent spoilage during sake brewing and to produce ginjoshu flavor. Accordingly, sake brewing in Akita improved significantly. A second key person was Uhei Sato V, the family head of Aramasa Brewery. Sake produced in Aramasa Brewery was awarded third place in 1927 and 1928 at the Annual Japan Sake Awards. These results led Fujio Oana, an engineer in the NRIB, to become interested in the Aramasa sake mash. Oana isolated a new strain of sake yeast in 1930 and began to distribute it in 1935 as Kyokai Strain No. 6. These three engineers – Masaaki Hanaoka, Uhei Sato V and Fujio Oana – all graduated from the Osaka High School of Engineering, which was the predecessor of the Faculty of Engineering, Osaka University. The birth of Kyokai Strain No. 6 can be attributed to collaborative efforts among these and other engineers.
Kyokai Strains No. 1–5 were isolated in Kansai and Hiroshima from the Meiji to the Taisho era. Kyokai Strain No. 6, and later strains, were isolated in other areas during the Showa era (). Although they had been distributed throughout the country, the old sake strains were probably not used in Akita. This may be why the new sake yeast strains emerged and were widely used in Japan later on.
Figure 3. Origin of representative sake yeast strains. (a) Prefectures where old yeast strains were isolated (Kyokai yeast strains No. 1–5), (b) Prefectures where new yeast strains were isolated (Kyokai yeast strains No. 6, No. 7, No. 9 and No. 10).
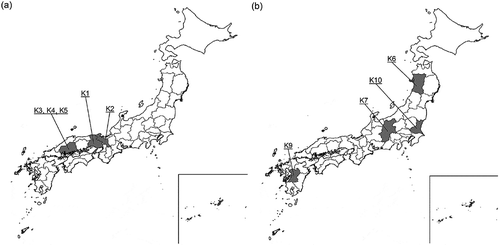
It is interesting to speculate why the older sake yeast strains were no longer distributed by the time of the Pacific War. Various plausible theories have been proposed; for example, that the characteristics of the old strains had changed unfavorably, or because new yeast strains brewed better sake. Although they are no longer widely distributed, the older sake yeast strains are of great traditional and historical importance and accordingly are preserved by the BSJ and the NRIB.
Breeding of various yeasts
Many sake yeasts have been developed for distribution by the BSJ and other contributors using four new yeast strains as parental strains (Kyokai Strains No. 6–10) () [Citation31]. Most commercial sake yeast strains are bred to form non-foaming cultures. Since bubbles form in the upper portion of the tank during sake fermentation, removing the upper layer of foam increases the practical utility of the tank as well as the improvement of workability by omitting bubble elimination work. Accordingly, many non-foam-forming yeasts (such as Kyokai Strains No. 601, No. 701, No. 901, No. 1001, No. 1401 and No. 1501) have been bred for mass sake production. Kyokai Strain No. 11 is resistant to high alcohol concentrations (> 18%), while Kyokai Strain No. 12 is suitable for ginjoshu production [Citation32–Citation35]. Kyokai Strains No. 1601, No. 1701 and No. 1801 have been bred to produce a large amount of isoamyl acetate and ethyl caproate (yielding a fruity aroma), and Kyokai Strain No. 1901 has been bred to be unable to produce urea [Citation36–Citation39]. This non-urea-producing yeast, with less ethyl carbamate, offers superior food safety and is more suitable for international distribution. The relationships among strains have been characterized (), which provides a basis for evaluating past breeding. Each prefecture’s brewery laboratory collects and maintains sake yeast independently, and diverse sake yeasts matching local tastes are also bred [Citation40].
Genetic variation among sake yeast strains
Recent DNA sequencing and large-scale population genomic surveys of 1,011 S. cerevisiae isolates revealed new genomic information about sake yeast strains [Citation25]. A ploidy study revealed that sake yeast strains are mostly diploids, but some are aneuploid. This is interesting because of the general mitotic growth advantage of euploid versus aneuploid strains. A DNA sequencing study revealed that sake yeast strains are closely related, and are very distinct from other populations, such as laboratory and wine yeast. The genetic variation among strains of sake yeast is typically low, although genetic variation within S. cerevisiae has been generally shaped by a complex history influenced by human activities such as domestication and admixture. It is widely-believed that S. cerevisiae originated from S. paradoxus, a closely related but distinct species within Saccharomyces sensu stricto. Several recent analyses support a Chinese origin for S. cerevisiae, and it appears that global S. cerevisiae strains descend from a single ancestor in China. Consistent with their geographical history, sake yeast strains diverged from Chinese isolates earlier than wine and beer yeast strains isolated in Europe. The timing of the S. cerevisiae – S. paradoxus speciation event, the out-of-China S. cerevisiae migration event, and the divergence dates for sake and wine yeast are tentatively estimated to be 300,000, 15,000, 4,000 and 1,500 years ago, respectively [Citation25]. One type of genetic event frequently observed in sake isolate genomes is loss of heterozygosity (LOH). LOH events represent up to 80% of sake isolate genomes. Sake isolates are characterized by low heterozygosity, suggesting a low outcrossing rate. All of these observations indicate that sake isolates experienced population expansion after a genetic bottleneck.
In-depth analysis of sake yeast neighbor-joining trees reveals that sake yeast strains are distributed in two clusters: the new strains and the old strains ()) [Citation24]. Since the new sake yeast strains were derived from the four related Kyokai strains, it is to be expected that the new strains cluster together. However, it is surprising the old sake yeast strains form a cluster. This suggests that nearly all of the yeast strains used in the Meiji and Taisho eras had a single origin.
Figure 4. Genetic and morphological cluster dendrogram of new and old sake yeast strains. The dendrogram was generated based on genetic similarity (a) and morphological similarity (b) computed by the correlation coefficient [Citation24]. Scale bar indicates the mean correlation coefficient. Orange and blue indicate new and old sake yeast strains, respectively.
![Figure 4. Genetic and morphological cluster dendrogram of new and old sake yeast strains. The dendrogram was generated based on genetic similarity (a) and morphological similarity (b) computed by the correlation coefficient [Citation24]. Scale bar indicates the mean correlation coefficient. Orange and blue indicate new and old sake yeast strains, respectively.](/cms/asset/010b3c62-8ddd-42ba-bee8-a1cb39114a98/tbbb_a_1564620_f0004_oc.jpg)
Morphological variation among sake yeast strains
Morphological analyses have been used to characterize sake yeast strains. In the Meiji era, Ryoji Nakazawa observed S. tokyo and S. yedo cells using light microscopy and found that they have distinct cell morphology [Citation9]. Both strains have round cells, but those of S. tokyo are much larger than those of S. yedo. Nakazawa compared these strains with S. sake and defined them as new species based on phenotypic differences. As described earlier, S. tokyo, S. yedo and S. sake subsequently turned out to be the same species, S. cerevisiae.
The recent development of CalMorph, a semiautomated image processing system, has allowed yeast morphology to be analyzed quantitatively and statistically. After manual staining of the yeast cell wall, actin and nuclear DNA, 501 morphological traits are automatically extracted by CalMorph and can be used for statistical analysis [Citation41,Citation42]. High-dimensional morphological analysis has shown that new sake yeast strains (Kyokai Strains No. 6, No. 7, No. 9 and No. 10) have distinct cell morphology compared with old sake yeast strains (S. sake, S. tokyo, S. edo; Kyokai Strains No. 1–5) [Citation24]. The new sake strains (orange dots) and the old strains (blue dots) were nearly completely separated in the morphologically degenerated space (). Clustering analysis was performed based on the phenotypic distance between sake yeast strains, and showed that the new sake strains and the old sake yeast strains clustered into two almost completely distinct groups ()). Cells of the old sake yeast strains are larger than those of the new sake yeast strains, and the old sake strains accumulate budded cells. This serves as a useful tool to distinguish new from old strains. Since the new sake yeast strains produce characteristically flavored sake, it is interesting to consider whether there is a direct relationship between morphology and fermentation properties.
Figure 5. Phenotypic distribution of K7-lineage strains and old sake yeast strains. Orange, blue and black circles indicate new sake yeast, old sake yeast and other strains, respectively. PC1 and PC2 represent the first and second principle components of the morphological space, respectively [Citation24]. Percentages in parentheses on each axis indicate the contribution ratio.
![Figure 5. Phenotypic distribution of K7-lineage strains and old sake yeast strains. Orange, blue and black circles indicate new sake yeast, old sake yeast and other strains, respectively. PC1 and PC2 represent the first and second principle components of the morphological space, respectively [Citation24]. Percentages in parentheses on each axis indicate the contribution ratio.](/cms/asset/3a6836c3-1323-4f3b-85db-a16dc9617aeb/tbbb_a_1564620_f0005_oc.jpg)
Application of high-dimensional phenotyping during the breeding and brewing processes
In addition to genomic analyses, high-dimensional morphological analyses have several applications in yeast breeding. First, morphological analysis serves as a tool to distinguish the new sake breeds from unfavorable, naturally emerging yeasts [Citation24]. Since morphological profiles are associated with genotypes, classification based on cell morphology using image processing provides opportunities to predict the geographic and ecological origins of a strain. To distinguish sake yeasts from other unfavorable yeasts, selection media containing β-alanine or 2,3,5-triphenyl tetrazolium chloride have been used [Citation43,Citation44]. Morphological examination may replace these methods if a system for monitoring yeast cell morphology can be introduced into sake brewery. Second, high-dimensional phenotyping is a useful tool for screening for off-target mutations accidentally incorporated during breeding. When a lineage map is known, the degree of morphological changes between parent and progeny can be individually evaluated. The use of mutagens results in substantial morphological alterations, possibly due to the accumulation of off-target mutations. One potential breeding strategy is to exclude undesirable segregants with large degrees of unexpected morphological change. Third, morphological features can be used to select for desired or undesired traits for sake fermentation. Our recent study indicated that morphological homogeneity can be used as a marker for breeding risk-free yeast [Citation37]. In other words, a significant increase in morphological variability could undermine robust fermentation. Therefore, morphological homogeneity is preferable during breeding. Identifying correlations between morphology and desired traits would enable new breeding strategies. Finally, combined with other omics studies, including metabolomics and transcriptomics, high-dimensional phenotyping of sake yeast strains has wider applications [Citation45,Citation46]. Correlation analyses among omics data may provide critical new knowledge for breeding. High-dimensional phenotyping generates new, and significantly more, data than previously possible. It provides a means of improving our understanding of breeding organisms and obtaining new knowledge for efficiently breeding new micro-organism varieties.
Acknowledgments
We thank Shota Nemoto and other members of the Laboratory of Signal Transduction for helpful discussions. This work was supported by the Japan Science and Technology Agency (15H04402 to Y.O.).
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Meyen J. Jahresbericht über die Resultate der Arbeiten im Felde der physiologischen Botanik von dem Jahre 1837. Arch Naturgesch. 1838;4:1–186.
- Hansen EC. Undersøgelser over Alkoholgjaersvampenes Fysiologi og Morfologi. II. Om Askosporedannelsen hos Slægten Saccharomyces. Meddelelser fra Carlsberg Laboratoriet. 1883;2:29–102.
- Pasteur L. MeÂmoire sur la fermentation alcoolique. Ann Chim Phys. 1860;58:323–426.
- Juhler JJ. Umbildung eines Aspergillus in eines Saccharomyceten (Vonl. Mitt.). Centralbl f Bakt II. 1895;1:16–17.
- Atkinson RW. The chemistry of saké-brewing. Memoirs Sci Dept Tokio Daigaku. 1881;6:1–73.
- Kosai Y. A study of the Ashio copper mountain mineral dentistry. Tokyo Kagakukai-si. 1892;13:143–200. Japanese.
- Kosai Y, Yabe K. Uber die bei der Sakebereitung beteiligten Pilze. Central F Bakt Abt II. 1895;1:619–620.
- Yabe K. On the origin of sake yeast (Saccharomyces Sake). Coll Agric Tokyo Bull. 1897;3:221–224.
- Nakazawa R. Zwei Saccharomyceten aus Sakehefe. Central F Bakt Abt II. 1909;22:529–540.
- Watanuki H, Hayashi K. Suitability for bread and wine making in natural yeast fermentation. Tokyo Kasei Gakuin Daigaku Kiyou. 2014;54:33–40. Japanese.
- Tukahara T. Taxonomic study of sake yeast (1). J Brew Soc Jpn. 1961;56:888–890. Japanese.
- Tukahara T. Taxonomic study of sake yeast (2). J Brew Soc Jpn. 1961;56:1003–1005. Japanese.
- Stelling-Dekker NM. Die sporogenen Hefen. In: Akade-mie van Wetenschappen te Amsterdam. 1931. p. 546.
- Lodder J, Kreger-Vanrij NJW. The yeasts: a taxonomic study. North-Holland Pub. Co; 1952. p. 713.
- Tukahara T. Taxonomic study of sake yeast (4). J Brew Soc Jpn. 1961;57:117–123. Japanese.
- Lodder J. The yeasts: a taxonomic study. 2nd rev. and enl ed. North-Holland Pub. Co; 1970. p. 1385.
- Schacherer J, Shapiro JA, Ruderfer DM, et al. Comprehensive polymorphism survey elucidates population structure of saccharomyces cerevisiae. Nature. 2009;458:342–345.
- Liti G, Carter DM, Moses AM, et al. Population genomics of domestic and wild yeasts. Nature. 2009;458:337–341.
- Akao T, Yashiro I, Hosoyama A, et al. Whole-genome sequencing of sake yeast Saccharomyces cerevisiae Kyokai no. 7. DNA Res. 2011;18:423–434.
- Akao T. Genomics of the sake yeasts. In: Kitamoto K, editor. Front Fermented Foods. Tokyo: CMC Pub. Co; 2015. p. 117–126. Japanese.
- Cromie GA, Hyma KE, Ludlow CL, et al. Genomic sequence diversity and population structure of Saccharomyces cerevisiae assessed by RAD-seq. G3 (Bethesda). 2013;3:2163–2171.
- Takahashi Y, Eda K, Okumura Z, et al. About varieties of sake yeast. Jozo Shikenjo Houkokusyo. 1914;54:1–66. Japanese.
- The Brewing Society of Japan. History of the 70th anniversary of The Brewing Society of Japan. 1975. p. 155. Japanese.
- Ohnuki S, Okada H, Friedrich A, et al. Phenotypic diagnosis of lineage and differentiation during sake yeast breeding. G3(Bethesda). 2017;7:2807–2820.
- Peter J, De Chiara M, Friedrich A, et al. Genome evolution across 1,011 Saccharomyces cerevisiae isolates. Nature. 2018;556:339–344.
- Azumi M, Goto-Yamamoto N. AFLP analysis of type strains and laboratory and industrial strains of Saccharomyces sensu stricto and its application to phonetic clustering. Yeast. 2001;18:1145–1154.
- Hanaoka M. Sake brewing essentials (5). Jozogaku Zassi. 1924;2:340–348. Japanese.
- Hanaoka M. Proposal of Akita style fermentation starter manufacturing process (1). Jozogaku Zassi. 1928;6:147–174. Japanese.
- Hanaoka M. Proposal of Akita style fermentation starter manufacturing process (2). Jozogaku Zassi. 1928;6:243–254. Japanese.
- Hanaoka M. Study on growth temperature of fermentation starter. J Brew Soc Jpn. 1934;29:12–16. Japanese.
- Kitagaki K, Kitamoto K. Breeding research on sake yeasts in Japan: history, recent technological advances, and future perspectives. Annu Rev Food Sci Technol. 2013;4:215–235.
- Ouchi K, Akiyama H. Non-foaming mutants of sake yeasts-selection by cell agglutination method and by froth flotation method. Agric Biol Chem. 1971;35:1024–1032.
- Shimoi H, Sakamoto K, Okuda M, et al. The AWA1 gene is required for the foam-forming phenotype and cell surface hydrophobicity of sake yeast. Appl Environ Microbiol. 2002;68:2018–2025.
- Hara M. Sake brewing with alcohol-tolerant yeast - New “Kyokai-yeast”. J Brew Soc Jpn. 1978;73:701–703. Japanese.
- Sato K. Kyokai Strain No. 12: the point of ginjo-shu brewing by Miyagi yeast. J Brew Soc Jpn. 1985;80:598–600. Japanese.
- Ichikawa E, Hosokawa N, Hata Y, et al. Breeding of a sake yeast with improved ethyl caproate productivity. Agric Biol Chem. 1991;55:2153–2154.
- Tamura H, Okada H, Kume K, et al. Isolation of a spontaneous cerulenin-resistant sake yeast with both high ethyl caproate-producing ability and normal checkpoint integrity. Biosci Biotechnol Biochem. 2015;79:1191–1199.
- Kitamoto K, Oda K, Gomi K, et al. Genetic engineering of a sake yeast producing no urea by successive disruption of arginase gene. Appl Environ Microbiol. 1991;57:301–306.
- Hasuda H. About urea non-productive high ester Kyokai Strain No.1901 (KArg 1901). J Brew Soc Jpn. 2014;109:576–581. Japanese.
- Ohya Y, Nemoto S. Sake yeast then and now: discovery of Saccharomyces sake and differentiated procedures for sake brewing. Jpn J Hist Brew. 2018;33:1–7. Japanese.
- Ohya Y, Sese J, Yukawa M, et al. High-dimensional and large-scale phenotyping of yeast mutants. Proc Natl Acad Sci USA. 2005;102:19015–19020.
- Ohya Y, Kimori Y, Okada H, et al. Single-cell phenomics in budding yeast. Mol Biol Cell. 2015;26:3920–3925.
- Sugama S, Yamakawa K, Kataoka G, et al. Study on sake yeast in sake breweries. J Brew Soc Jpn. 1965;60:453–456. Japanese.
- Nakamura T. Management of microorganisms. J Brew Soc Jpn. 1998;93:586–593. Japanese.
- Mimura N, Isogai A, Iwashita K, et al. Gas chromatography/mass spectrometry based component profiling and quality prediction for Japanese sake. Biosci Bioeng. 2014;118:406–414.
- Hirasawa T, Furusawa C, Shimizu H. Saccharomyces cerevisiae and DNA microarray analyses: what did we learn from it for a better understanding and exploitation of yeast biotechnology? Appl Microbiol Biotechnol. 2010;87:391–400.
- Akao T. New perspectives of genetic variability of sake yeast strains in microevolution and breeding. Kagaku Seibutu. 2014;52:223–232. Japanese.
