ABSTRACT
Glycyrrhiza glabra is considered as potential drug for nasopharyngeal carcinoma (NPC). However, whether the long noncoding RNAs’ (lncRNAs) contributes to the anti-cancer function of this herb is unknown. In present study, we analyzed the differential expression of lncRNA between G. glabra-treated and untreated C666-1 cells. Out of those tumor-related lncRNAs, AK027294 had a strongest down-regulation upon G. glabra treatment. Knockdown of AK027294 suppresses the proliferation of C666-1 cells by inducing the apoptosis. Moreover, either G. glabra treatment or knockdown of AK027294 significantly increases the production of EZH1 (Enhancer of zeste 1 polycomb repressive complex 2 subunit). Collectively, we have identified a potential mechanism that the down-regulation of AK027294 contributes to the anti-cancer function of G. glabra and also provide the potential inter-relationship between AK027294 and EZH1.
Graphical abstract

One lncRNA, AK027294 contributes to the anti-cancer activity of G. glabra in nasopharyngeal carcinoma cell line, C666-1.
KEYWORDS:
In southern China and Southeast Asia, nasopharyngeal carcinoma (NPC) has an exceptionally high incidence and is prone to relapse in many clinical patients, although most NPCs are undifferentiated and strongly radiosensitive [Citation1–Citation3]. A line of studies reveal that traditional Chinese medicine (TCM) may serve as latent drugs for the therapeutic intervention of cancer [Citation4,Citation5]. TCM is capable of modulating the immune system and minimizing the adverse effects of cancer treatment, indicating that TCM is efficacious as a concomitant therapy for NPC patients [Citation6,Citation7]. Glycyrrhiza glabra, a common herbal drug in Western and Eastern medicine for centuries, has been currently employed for treating inflammation, allergy and some kind of cancers [Citation8,Citation9]. Recently, the anti-cancer activity of G. glabra on NPC has been reported [Citation10].
Recent studies have additionally revealed that the human genome is transcribed to produce almost 15,000 long non-coding RNAs (lncRNAs) [Citation11], which play crucial roles in a wide range of cellular processes such as apoptosis, cell cycle, migration, and invasion [Citation12–Citation15]. Over the last decade, there were many reports of aberrant expression of lncRNAs in hepatocellular carcinoma, lung cancer, and breast cancer [Citation16–Citation18], which revealed that dysregulated lncRNAs were closely related to human cancer and function as tumor suppressors or oncogenes [Citation19,Citation20]. In NPC, a novel lncRNA n375709 was identified by next generation deep sequencing, which was overexpressed and led to paclitaxel resistance [Citation21]. A recent study evidenced that ANRIL was up-regulated in NPC cell lines and NPC tissues and promoted NPC progression through activating cell proliferation, reprograming cell glucose metabolism and inducing side-population stem-like cancer cells [Citation22]. Undoubtedly, lncRNAs have become new players close related to cancer, although the most pathophysiological contributions and response mechanisms of lncRNAs to human NPC remains limited. Additionally, recent studies have indicated that, the mechanisms of anti-cancer effects by lncRNA could be achieved by regulating the expression of some key oncogenes or tumor suppressor genes and affect the occurrence and development of tumor through lncRNA-miRNA or lncRNA-mRNA interaction [Citation23].
In this work, we overviewed the aberrant expression of G. glabra-treated lncRNAs in NPC cell line, C666-1, focusing on the effect of the lncRNA on the proliferation of NPC, and investigating the mechanisms and functions of lncRNAs on NPC and their potentials as biomarkers and targets for novel therapeutic approaches in the future.
Materials and methods
Extraction of water extract of G. glabra
The extraction of water extract of G. glabra was performed as previously described [Citation10]. Briefly, 400 g of G. glabra roots were extracted with purified water (400 mL) using a reflux for 3h at 100°C and then filtered using a 25 μm sieve. The extracts were lyophilized into ~29.2 g dry powder, which was kept in a deep freezer (−80 oC). For experiments, 100 mg dry powder was re-dissolved in 100 mL RPMI-1640 culture medium (HyClone), forced through a 0.45 μm syringe filter before use.
Cell culture and viability assay
Human nasopharyngeal carcinoma cell line, C666-1 was cultured in RPMI-1640 culture medium (Hyclone) supplemented with 10% (v/v) FBS and 100 units penicillin/streptomycin at 37°C with 5% CO2 atmosphere in a humidified incubator. Cells grown in flask were allowed approximately to reach 90% confluence. The culture medium was renewed every two day for better growth of cells and then were rinsed and removed from the flask by incubating them with a trypsin-EDTA solution (Hyclone), and harvested in a 15 mL centrifuge tube for subsequent study.
To determine the anti-cancer of G. glabra root extract in C666-1, cells were seeded and grown to 80 – 90% confluence prior to treatment with this extract, at increasing concentration (0.25, 0.5, and 1 mg/mL). After 48 hours, cells were collected for the MTT viability analysis. Briefly, 2 mg/mL of MTT solution was added to each well and incubated for 3 h at 37°C. The supernatant was discarded and the blue formazan crystals were dissolved in 200 μL of DMSO and 25 μL Sorenson buffer. The absorbance was read in a microplate reader (Biotek) at 570 nm. Each experiment was repeated in a triplicate.
LncRNA profiling
To identify the involved lncRNAs, Human Disease-related LncRNA Profiler (System Biosciences) was used and overall lncRNAs were selected from either the RNA database (http://research.imb.uq.edu.au/rnadb/Default.aspx) or lncRNA database (www.lncRNAdb.org). Total RNA was extracted from G. glabra-treated and untreated C666-1 cells. Reverse transcription was performed by using RevertAidTM Reverse Transcriptase (Fermentas) and random primer mix (New England BioLabs). The values for the cells without treatment after normalization by the internal controls served as a basal level of expression of indicated lncRNAs; delta-delta Ct values (no treatment versus G. glabra treatment) were used to determine their relative expression as fold changes.
RNA interference
Basing on the sequence of AK027294, three small interference RNA (siRNA #1: 5ʹ-ttacttggtgccaagcacta-3ʹ; siRNA #2: 5ʹ-ctttgctgaggaaatttaca-3ʹ, and siRNA #3: 5ʹ-attgtctataatctcgctag-3ʹ) were synthesized (Qiagen) and negative control, scrambled siRNA, were purchased from Ambion. C666-1 cells were transfected with siRNA (200nM) and corresponding Lipofectamine 2000 (Invitrogen) according to the manufacture’s instruction. After incubated for 24 hours, cells were collected for follow experiments.
Cell proliferation assay
The cells were seeded in 96-well plates 24 h prior to transfection, then siRNAs were used to transfect the cells. The transfected cells were collected at different time points (12, 24, 36 and 48 hour). Cell counting kit-8 (CCK-8, Dojindo) was used to measure the cell proliferation following the manufacturer’s instruction.100μL CCK-8 was added and incubated at 37°Cfor 3h, then the number of cells was measured at a wavelength of 450 nm. All the experiments were carried out in triplicate.
Flow cytometry analysis of apoptosis
The apoptosis of C666-1 was examined using dual-labeling with an annexin V-FITC and PI, fluorescence intensity was detected by flow cytometry. Briefly, cells with 80 – 90% confluence were transfected by siRNA. After 24 hours, the siRNA-transfected cells were collected, washed twice with PBS and suspended in 100 uL buffer (10mM HEPES/NaOH, pH 7.4, 140mM NaCl, 2.5mM CaCl2). Subsequently, 5 µL Annexin V-FITC were added and incubated at room temperature in the dark for 10 min and then cells washed in PBS and 10 µL of PI was loaded. Finally, the percentage of apoptosis cells was determined by flow cytometry at a wavelength of 515 nm for FITC and 560nm for PI. All the experiments were carried out in triplicate. Data analysis was performed with a BD BioSciences FACSCalibur flow cytometer using CellQuest sofware.
Western blotting
Protein levels of Caspase 8, Caspase 3, Caspase 9 and EZH1 were examined by western blot analysis. Transfected or G. glabra treated cells were collected, and lysed in lysis buffer containing 20mM Tris (pH 8.0), 137mM EDTA, 100mM NaF, 1mM phenylmethylsulfonyl fluoride, 0.25 trypsin inhibitory units/mL aprotinin and 10mg/mL leupeptin. 50µL of total proteins were separated using 10% SDS-polyacrylamide gels and then transferred to polyvinylidene difluoride membranes. Membranes were blocked with 5% nonfat milk and then probed with a primary antibodies against human Caspase 8, Caspase3, Caspase9, EZH1 and Glyceraldehyde-3-phosphate dehydro-genase (GAPDH), followed by secondary antibody conjugated with horseradish peroxidase. Bound antibodies were detected using the enhanced chemi luminescence (ECL) kit (Thermo scientific). All the experiments were carried out in triplicate.
Quantitative RT-PCR assay
Total RNA of transfected cells was extracted witha TRIzol kit (Invitrogen) and converted to cDNA with a first strand cDNA synthesis kit (Fermentas). For the detection of AK027294 and EZH1 expression, the following primer sequences were used for RT-PCR analysis: AK027294: 5ʹ-ATGACACCTATTGGAGAA-3ʹ (sense), 5ʹ-TAAGCACACCTGAGTAAT-3ʹ (anti-sense); EZH1: 5ʹ- GAGTTGGTCGATGCCCTGAAT-3ʹ (sense), 5ʹ- AGCATGTCGCTTTCTCTTTC TT-3ʹ (anti-sense); GAPDH: 5ʹ-CTCACCGGATGCACCAATGTT-3ʹ (sense), 5ʹ- CGCGTTGCTCACAATGTTCAT-3ʹ (anti-sense), and GAPDH acted as a reference gene to normalize mRNA concentrations. Finally, amplification and detection of gene expression levels were analyzed by 7500 real-time PCR System (Applied Biosystems).
Statistical analysis
The unpaired two-tailed Student t test were used to analyze the statistical significance of cell viability, cell proliferation, and mRNA levels. All data were analyzed by Prism (GraphPad Software, Inc.), and P values less than 0.05 and 0.01 were deemed as two significant levels.
Results
LncRNAs are expressed in tumor and non-tumor pancreatic tissues
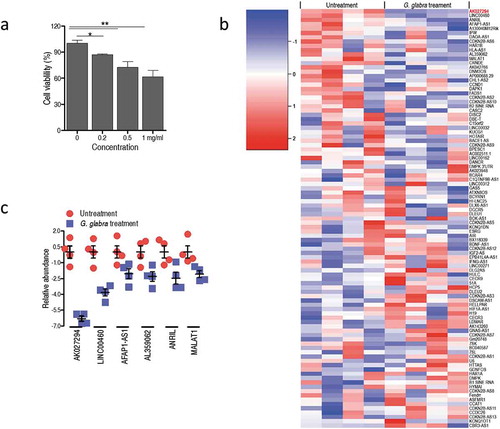
To confirm the anti-cancer effect of G. glabra root extract on the growth of C666-1 cells, MTT assay was used to measure the proliferation of C666-1 cells treated by various concentrations of the drug. As anticipated, the G. glabra root extract obviously inhibits C666-1 cell growth in a dose-dependent manner ()). Previous study showed that aberrant expression of lncRNAs was closely related to human NPC [Citation21,Citation22]. Interestingly, our lncRNA profiling showed that the treatment of G. glabra root extract is able to alter the expression of tumor-related lncRNAs ()). For above tumor-related lncRNAs, generally, abundance was differentially increased in the cells that avail tumorigenesis (or vice versa). Given that, we primarily focus on the lncRNAs which were down-regulated by G. glabra root extract. Among the eight down-regulated lncRNA in C666-1 cells, AK027294 presented the highest fold change ()).
Knockdown of AK027294 has cell proliferation suppression effects
To examine the function of AK027294 to the proliferation of C666-1 cells, we knocked down the expression level of AK027294 using RNA interference (RNAi) technique. Three siRNAs that target AK027294 were constructed, and their effects were analyzed by transfection into C666-1 cells. Compared to C666-1 cells with scrambled siRNA control, the expression levels of AK027294 was significantly decreased in the AK027294 siRNA-transfected cells ()), demonstrating the effectiveness of AK027294 siRNA. Specifically, the ability to decrease the expression of AK027294 show a hierarchy of siRNA #2 > siRNA #3 > siRNA #1.
Figure 2. Knockdown of AK027294 inhibits the proliferation of C666-1 cells through inducing the apoptosis.
(a) Three AK027294 siRNAs decrease the expression of AK027294 in C666-1 cells. (b) AK027294 silencing inhibits the proliferation of C666-1 cells. (c) AK027294 silencing induces the apoptosis of C666-1 cells. (d) AK027294 silencing increases the translational level of caspase 3, 8, and 9 in C666-1 cells. Error bars ± SEM, * P < 0.05 and ** P < 0.01
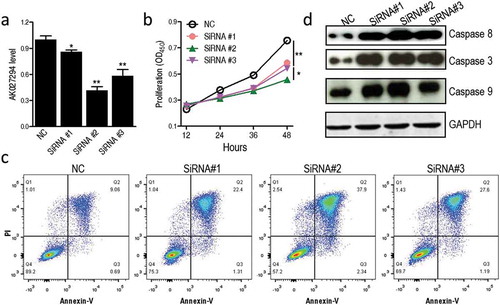
Next, cell proliferation was measured by using CCK-8 assay in C666-1 cells transfected with AK027294 siRNAs and scrambled siRNA. Interestingly, AK027294 siRNA-transfected cells have a lower proliferation in contrast to scrambled siRNA-transfected cells ()), indicating that the knockdown of AK027294 inhibits the proliferation of C666-1 cells. Of note, the proliferation of C666-1 is positively correlated with the expression level of AK027294. To further examine whether AK027294 silencing inhibits the proliferation of C666-1 cells through inducing apoptosis, the apoptosis of transfected cells was determined by flow cytometry using annexin V-FITC and PI. In contrast to scrambled siRNA-transfected cells, the AK027294 siRNA-transfected cells have an obviously higher apoptosis ()). Meanwhile, the activity of caspase-3, caspase-9, and caspase-8 were examined by western blotting, as activation of caspase cascades has been shown to occur in apoptosis [Citation24]. Compared with scrambled siRNA-transfected cells, the increase protein level of caspase-3, caspase-8, and caspase-9 were detected in cells incubated with AK027294 siRNAs ()).
Knockdown of AK027294 leads to increase EZH1 expression
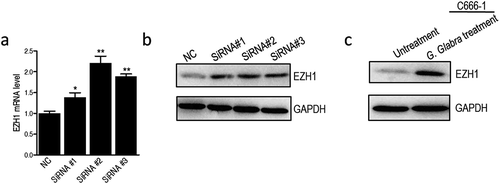
Previous study already showed that lncRNA was involved in the tumorigenesis through the regulation of EZH1 [Citation25]. To understand whether the cell proliferation suppression by AK027294 silencing was linked to the EZH1, we measured the EZH1 expression in AK027294 silencing C666-1 cells from mRNA and protein levels. Surprisingly, C666-1 Cells with AK027294 siRNAs showed the significantly higher mRNA levels of EZH1 than scrambled siRNA-transfected cells ()). This finding suggested that AK027294 silencing have an ability to improve the EZH1 expression in C666-1 cells. We also examine the protein levels of EZH1. As ) showed, the protein levels of EZH1 were notably increased in C666-1 cells transfected with AK027294 siRNAs compared to scrambled siRNA-transfected cells. Furthermore, the treatment of G. glabra root extract was also capable of augmenting the production of EZH1 in C666-1 cells ()), thereby providing a potential mechanism that AK027294 silencing suppresses C666-1 proliferation by increasing EZH1 expression levels.
Figure 3. AK027294 silencing boosts the expression of EZH1 in C666-1 cells.
(a) Transcriptional level of EZH1 is boosted by AK027294 siRNAs. (b) AK027294 silencing increases the production of EZH1 in C666-1 cells. (c) G. glabra-treated C666-1 cells show the boosted production of EZH1. Error bars ± SEM, * P < 0.05 and ** P < 0.01
Discussion
Recently, G. glabra has been considered as a potential drug against NPC [Citation10]. However, the underlying mechanisms by which this herb induces anti-cancer activity is largely unknown. A line of evidences has indicated lncRNAs closely linked to NPC, and the differential expression of lncRNAs was identified between tumor and non-tumor tissue samples [Citation21,Citation22], but the potential function and the molecular mechanism of lncRNAs in the pathogenesis of NPC are much less understood [Citation26]. In present study, we investigated the inter-relationship between G. glabra treatment and lncRNA, and in follow-up experiments, we researched its function and potential molecular mechanism of C666-1 cell proliferation.
The core discovery in our study is that several differential lncRNAs are identified between untreated and G. glabra treated samples, and the most significantly down-regulated lncRNA (AK027294) upon G. glabra treatment was chosen to identify its function and potential molecular mechanisms in NPC proliferation. Although the previous studies had demonstrated that AK027294 was closely related to breast, gastric, lung and colorectal cancer [Citation27–Citation30], the detail functions and its molecular mechanisms in NPC proliferation was much less understood. Currently, we present that AK027294 silencing resulted in proliferation arrest by increasing apoptosis. As the propensity for early metastatic of NPC [Citation31,Citation32], that AK027294 silencing induce inhibition of cell proliferation may be a potential approach to treat NPC. The published data indicate four dominant constituents of the water extract of G. glabra, which are Benzeneacetic acid, 4-hydroxy-, methyl ester (27.35%), Thiophene, Tetrahydro-2-methyl- (11.42%), Mome-Inositol (9.91%), and 5-Tridecanone (4.73%) [Citation8]. In future research, we would address which compound is/are responsible for the regulation of AK027294 in NPC cells.
In mammalian, PRC2 complexes, including Ezh1 and Ezh2, contain chromatin-modifying histone lysine methyl-transferases [Citation33]. Ezh2 catalyzes the trimethylation of histone-tail H3-lysine 27 residues associated with embryonic development [Citation34,Citation35]. Delineation of the function of Ezh1 has been more challenging as some studies suggested that EZH1 presented a weakly activity of H3K27 methyl-transferase and mentioned as a complement to EZH2 [Citation36]. In contrast, recent studies in developing hippocampal neurons concluded that EZH1 acted in an antagonistic role of EZH2 [Citation35]. Besides, EZH2 was initially found to be elevated in a subset of cancers, almost all metastatic prostate cancers, breast cancer, melanoma, bladder cancer and gastric cancer [Citation37–Citation41]. At present, there is no reports has shown the function of EZH1 related to cancer, but in repopulating HSCs, EZH1 suppresses a cell-cycle and prevents differentiation [Citation42]. In our study, we present that AK027294 silencing notably up-regulated the expression of EZH1, and resulted in proliferation arrest. Collectively, we presume that AK027294 silencing suppresses the proliferation of NPC by inducing the apoptosis is based on up-regulating EZH1 expression levels. Certainly, we would determine the mechanism of AK027294 regulating EZH1 expression in future study.
Taken all together, we demonstrate a plausible lncRNA-regulated mechanism that AK027294 is responsible for the anti-cancer activity of G. glabra. Besides, either AK027294 silencing or G. glabra treatment induces the expression of EZH1, which provides the possible mechanistic link for the downstream mechanism of AK027294.
Author contribution
Bo Zhang, Wei Zhang, Zhao-yang Ke, and Ling-guo Ma performed experiments. All authors were responsible for the data analysis and figure format. Bo Zhang and Min Yan participated in paper writing. Min Yan was responsible for the research design and modifying the manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- De Martel C, Ferlay J, Franceschi S, et al. Global burden of cancers attributable to infections in 2008: a review and synthetic analysis. Lancet Oncol. 2012;13:607–615.
- Cho WC, Chen H. Clinical efficacy of traditional Chinese medicine as a concomitant therapy for nasopharyngeal carcinoma: a systematic review and meta-analysis. Cancer Invest. 2009;27:334–344.
- Yin SY, Wei WC, Jian FY, et al. Therapeutic applications of herbal medicines for cancer patients. Evid Based Complement Alternat Med. 2013;2013:302426.
- Yanwei L, Yinli Y, Pan Z. Traditional herbal formula NPC01 exerts antiangiogenic effects through inhibiting the PI3K/Akt/mTOR signaling pathway in nasopharyngeal carcinoma cells. Evid Based Complement Alternat Med. 2018;2018:5291517.
- Zhao M, Luo CM, Long F, et al. Growth capability of epithelial cell line of human poorly differentiated nasopharyngeal carcinoma and its response to Chinese medicinal herbs and marine drugs. Zhonghua Zhong Liu Za Zhi. 1988;10:98–101.
- Kim W, Lee WB, Lee J, et al. Traditional herbal medicine as adjunctive therapy for nasopharyngeal cancer: a systematic review and meta-analysis. Integr Cancer Ther. 2015;14:212–220.
- Chien CR, Su SY, Cohen L, et al. Use of Chinese medicine among survivors of nasopharyngeal carcinoma in Taiwan: a population-based study. Integr Cancer Ther. 2012;11:221–231.
- Thiyagarajan P, Chandrasekaran CV, Deepak HB, et al. Modulation of lipopolysaccharide-induced pro-inflammatory mediators by an extract of Glycyrrhiza glabra and its phytoconstituents. Inflammopharmacology. 2017;3:47–53.
- Nourazarian SM, Nourazarian A, Majidinia M, et al. Effect of root extracts of medicinal herb Glycyrrhiza glabra on HSP90 gene expression and apoptosis in the HT-29 colon cancer cell line. Asian Pac J Cancer Prev. 2016;16:8563–8566.
- Zheng C, Han L, Wu S. A metabolic investigation of anticancer effect of G. glabra root extract on nasopharyngeal carcinoma cell line, C666-1. Mol Biol Rep. 2019;46:1–8.
- Derrien T, Johnson R, Bussotti G, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789.
- Khaitan D, Dinger ME, Mazar J, et al. The melanoma-upregulated long noncoding RNA SPRY4-IT1 modulates apoptosis and invasion. Cancer Res. 2011;71:3852–3862.
- Meola N, Pizzo M, Alfano G, et al. The long noncoding RNA Vax2os1 controls the cell cycle progression of photoreceptor progenitors in the mouse retina. RNA. 2012;18:111–123.
- Gupta RA, Shah N, Wang KC, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076.
- Tang LH, Zhang W, Su B, et al. Long noncoding RNA HOTAIR is associated with motility, invasion, and metastatic potential of metastatic melanoma. Biomed Res Int. 2013;2013:251098.
- Braconi C, Kogure T, Valeri N, et al. microRNA-29 can regulate expression of the long non-coding RNA gene MEG3 in hepatocellular cancer. Oncogene. 2011;30:4750–4756.
- Yang Y, Li H, Hou S, et al. The noncoding RNA expression profile and the effect of lncRNA AK126698 on cisplatin resistance in non-small-cell lung cancer cell. Plos One. 2013;8:e65309.
- Augoff K, McCue B, Plow EF, et al. miR-31 and its host gene lncRNA LOC554202 are regulated by promoter hypermethylation in triple-negative breast cancer. Mol Cancer. 2012;11:5.
- Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1:391–407.
- Qi P, Du X. The long non-coding RNAs, a new cancer diagnostic and therapeutic gold mine. Mod Pathol. 2013;26:155–165.
- Ren S, Li G, Liu C, et al. Next generation deep sequencing identified a novel lncRNA n375709 associated with paclitaxel resistance in nasopharyngeal carcinoma. Oncol Rep. 2016;36:1861–1867.
- Zou ZW, Ma C, Medoro L, et al. LncRNA ANRIL is up-regulated in nasopharyngeal carcinoma and promotes the cancer progression via increasing proliferation, reprograming cell glucose metabolism and inducing side-population stem-like cancer cells. Oncotarget. 2016;7:61741.
- Hu G, Niu F, Humburg BA, et al. Molecular mechanisms of long noncoding RNAs and their role in disease pathogenesis. Oncotarget. 2018;9:18648.
- Danial NN, Korsmeyer SJ. Cell death: critical control points. Cell. 2004;116:205–219.
- Sun Y, Zhou Y, Bai Y, et al. A long non-coding RNA HOTTIP expression is associated with disease progression and predicts outcome in small cell lung cancer patients. Mol Cancer. 2017;16:162.
- Rinn JL, Kertesz M, Wang JK, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323.
- Feng YM, Li X, Sun B, et al. Evidence for a transcriptional signature of breast cancer. Breast Cancer Res Treat. 2010;122:65–75.
- Niu H, Hu Z, Liu H, et al. Long non-coding RNA AK027294 involves in the process of proliferation, migration, and apoptosis of colorectal cancer cells. Tumour Biol. 2016;37:10097–10105.
- He X, Pan XM, Jin MM, et al. Long non-coding RNA AK027294 promotes tumor growth by upregulating PCNA in gastric cancer. Eur Rev Med Pharmacol Sci. 2019;23:5762–5769.
- Chen B, Ling C. Long noncoding RNA AK027294 acts as an oncogene in non-small cell lung cancer by up-regulating STAT3. Eur Rev Med Pharmacol Sci. 2019;23:1102–1107.
- Khanfir A, Frikha M, Ghorbel A, et al. Prognostic factors in metastatic nasopharyngeal carcinoma. Cancer Radiother. 2007;11:461–464.
- Su SF, Han F, Zhao C, et al. Long-term outcomes of early-stage nasopharyngeal carcinoma patients treated with intensity-modulated radiotherapy alone. Int J Radiat Oncol Biol Phys. 2012;82:327–333.
- Margueron R, Reinberg D. The polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349.
- Su IH, Basavaraj A, Krutchinsky AN, et al. Ezh2 controls B cell development through histone H3 methylation and Igh rearrangement. Nat Immunol. 2003;4:124–131.
- Henriquez B, Bustos FJ, Aguilar R, et al. Ezh1 and Ezh2 differentially regulate PSD-95 gene transcription in developing hippocampal neurons. Mol Cell Neurosci. 2013;57:130–143.
- Shen XH, Liu Y, Hsu YJ, et al. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol Cell. 2008;32:491–502.
- Varambally S, Dhanasekaran SM, Zhou M, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629.
- Kleer CG, Cao Q, Varambally S, et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci U S A. 2003;100:11606–11611.
- Bachmann IM, Halvorsen OJ, Collett K, et al. EZH2 expression is associated with high proliferation rate and aggressive tumor subgroups in cutaneous melanoma and cancers of the endometrium, prostate, and breast. J Clin Oncol. 2006;24:268–273.
- Varambally S, Cao Q, Mani RS, et al. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science. 2008;322:1695–1699.
- Matsukawa Y, Semba S, Kato H, et al. Expression of the enhancer of zeste homolog 2 is correlated with poor prognosis in human gastric cancer. Cancer Sci. 2006;97:484–491.
- Hidalgo I, Herrera-Merchan A, Ligos JM, et al. Ezh1 is required for hematopoietic stem cell maintenance and prevents senescence-like cell cycle arrest. Cell Stem Cell. 2012;11:649–662.