ABSTRACT
This study aimed to explore the influence of Tryptophanyl-tRNA synthetase (WARS) expression on the proliferation and migration of uveal melanoma (UM) cells, and the potential mechanisms. Bioinformatics analysis based on Gene Expression Omnibus (GEO) database showed that WARS expression in metastatic cancer was significantly higher than that in no-metastatic group. Kaplan-Meier analysis based on The Cancer Genome Atlas (TCGA) database showed that high WARS expression was associated with lower survival. Biological function experiments showed that overexpression of WARS in OCM-1A cells can promote cell proliferation, migration, and invasion, whereas knockdown of WARS in C918 cells showed the opposite effect. Finally, we observed that the up-regulation of WARS induced the activation of phosphatidylinositol 3-kinase/AKT (PI3K/AKT) signaling, whilst depletion of WARS resulted in opponent outcomes. Taken together, our results illustrated that WARS was overexpressed in UM cells and contributed to the viability and motility of UM cells via modulating PI3K/AKT signaling pathway.
Graphical Abstract
Knockdown of WARS suppresses UM cell proliferation, migration, and invasion
Uveal melanoma (UM), as a rare intraocular malignant tumor, is common in Caucasians [Citation1]. It is easily misdiagnosed because of its complete asymptomatic early stage, and it is usually diagnosed at an advanced stage [Citation2]. Although local tumors can be well controlled at present, the high metastasis rate leads to poor overall therapeutic effect [Citation3]. Despite the progress of treatment technology, the improvement of five-year survival rate was still limited [Citation4]. Previous studies have shown that metastasis was a major cause of untreatable and mortality in UM patients [Citation5],and some clinical micrometastases are difficult to detect in diagnosis [Citation6]. Once metastases are found, there is no effective treatment [Citation7]. Hence, it is particularly important to study a gene that abnormally expressed in metastatic UM, which can help us find a new suitable treatment for UM patients.
Tryptophanyl-tRNA synthetase (WARS) is a member of the aminoacyl-tRNA synthase family [Citation8], also known as TRPRS, WRS, which is a potential prognostic marker of metastasis [Citation9]. Aminoacyl-tRNA synthase has been shown to play a mediating role in human tumorigenesis and cell metabolism. WARS, as one of them, has been found to be an imbalance in a variety of cancers [Citation10–Citation14], such as oral cancer, ovarian cancer, pancreatic cancer, colorectal cancer, etc. Previous studies have reported that WARS functions in a variety of physiological and pathological processes [Citation15–Citation19]. For example, it was participated in the immune response in guinea pigs with delayed hypersensitivity and Drosophila. Some studies have confirmed that WARS is associated with human angiogenesis. However, no studies have reported whether it plays a role in UM. Through bioinformatics analysis, we identified that WARS was also imbalanced in UM, hinting that regulating the expression of WARS may assist in the treatment for UM. Hence, based on the published literatures and bioinformatics analysis results, we chose it for further study.
As an important signal transduction pathway, PI3K/AKT has been widely reported to contribute to the occurrence and development of various cancers in recent years [Citation20]. Several studies have indicated that the phosphatidylinositol 3-kinase/AKT/mammalian target of rapamycin (PI3K/AKT/mTOR) pathway was closely related to the pathogenesis of UM. Ye et al. demonstrated that the PI3K/AKT signaling pathway has participated in UM cell migration [Citation21]. In addition, it has been reported that activated PI3K/AKT attenuates the inhibitory effect of rapamycin on UM cell proliferation [Citation22]. Nevertheless, whether WARS involve in the occurrence and development of UM through PI3K/AKT pathway remains to be clarified.
The purpose of this study was to confirm the correlation between the expression of WARS and clinical characteristics and survival rate of UM patients, and to investigate whether the regulation of WARS expression level would affect the invasion, migration, proliferation of UM cells, as well as the regulatory pathways, expecting to provide a new possible idea for UM clinical treatment.
Materials and methods
Data collection
We downloaded the data of 35 metastasis UM samples and 28 no-metastasis UM samples from the Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/) (access number: GSE22138), and the differential gene analysis was performed by GEO online analysis tool GEO2R. This study used RNA-Seq expression data and clinical data of UM from The Cancer Genome Atlas (TCGA) database (https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga) to analyze the correlation between WARS expression and the prognosis of UM patients.
Cells culture
We obtained human UM cell lines (OCM-1A, MUM-2C, C918, and MUM-2B) and normal control cells (D78) from the Shanghai Cell Bank of the Chinese Academy of Medical Sciences. DMEM medium, which containing FBS (10%), penicillin (100U/mL) and streptomycin (0.1mg/mL), was used for conventional culture at 37℃ with 5% CO2.
Cell transfection
The WARS siRNAs and nonspecific siRNA (si-con) were synthesized from Shanghai GenePharma Co., Ltd. (Shanghai, China) (si-WARS#1:TTTGACATCAACAAGACTT; si-WARS#2: CAGCACCTACCAGTAATCA; si-con: AATTCTCCGAACGTGTCACGT). UM cells were inoculated into 6-well plate 24 h before transfection and cultured in antibiotic-free medium. When cell confluence reached 80%, using Lipofectamine 2000 for transfection according to the instructions of the supplier. The transfected cells were incubated with 5% CO2 at 37℃for 6 h, and then the antibiotic-free medium was replaced by the complete culture medium. After 48 h, the expression of the transfected gene could be observed.
RNA extraction and quantitative real-time PCR (qRT-PCR)
We performed a qRT-PCR assay to test the mRNA expression level of WARS. Total RNA was extracted by TRIzol (Invitrogen), and SYBR Premix Ex Taq II (TaKaRa, Japan) were used for reverse transcription, which was executed according to the manufacturer’s agreement. Then, the expression of WARS was quantified by qRT-PCR, and the conditions were set with reference to the qRT-PCR kit. The primer sequences were as follows: WARS F:5ʹ- ATCAGAAACATGCCCTGGGG-3ʹ, R: 5ʹ- TTCAGTTGAGCACCTCCGAC-3ʹ; GAPDH:F:5ʹ-TGTGTCCGTCGTGGATCTGA-3ʹ, R: 5ʹ- CCTGCTTCACCACCTTCTTGA-3ʹ. Finally, the expression of WARS was calculated by 2−△△Ct method. Repeat the experiment three times independently.
Western blotting assay
RIPA lysate (containing protease inhibitor) was applied for cracking cells to extract protein, and BCA kit was used to detect protein concentration. Add about 20 μg of protein to each well in the vertical electrophoresis tank, and electrophoresis with 10% SDS page gel. The protein on the gel was transferred to PVDF membrane and blocked with 5% defatted milk powder for 1 h. Then, the primary antibodies were added and incubated overnight at 4℃. Primary antibodies used in our study were included as follows: anti-WARS (Abcam, Cambridge, UK); anti-AKT (1:500, Abcam, Cambridge, UK), anti-p-AKT (Ser473) (1:500, Abcam, Cambridge, UK), anti-mTOR (dilution 1:500, Abcam, Cambridge, UK), anti-p-mTOR(Ser2448) (1:1000, Abcam, Cambridge, UK), and anti-GAPDH (G5262, Sigma-Aldrich, USA). The next day, after a third wash with TBST, incubated the membrane at room temperature for 1 h with HRP-conjugated secondary antibody (sc-516,102, Santa Cruz Biotechnology, Inc., USA). Afterward, ECL developer was added after rinsing. Using GAPDH as a control to assess the relative protein levels.
CCK8 and colony formation assay
For CCK8 assay, cells were inoculated into 96-well plates with about 1000 cells per well and cultured in a carbon dioxide incubator. Cell viability was detected at 24 h, 48 h, and 72 h, according to the instructions of the CCK-8 kit (Dojindo Molecular Technologies, Rockville, MD, USA). The absorbance was detected at 450 nm using a microplate reader (Bio-Rad, CA, USA).
Simultaneously, we carried out a colony formation assay to test the colony formation ability of UM cells. About 400 cells were inoculated in culture dish which contains 5 mL pre-heated medium. Culture the cells for 1–2 weeks in a cell incubator at 37℃ with 5% CO2 and saturated humidity until naked-eye-visible clones appeared. The cells were fixed with 4% paraformaldehyde 5 mL for 30 m. Then, remove the fixative and dye the cells with 0.1% crystal violet dye for 30 min. Finally, number of colonies were counted.
Transwell assay
In invasion assay, Matrigel matrix glue 100μL (serum-free medium diluted at 1:6, BD Biosciences, NY, USA.) was added to the upper chamber of Transwell chamber (Chemicon International, Temecula, CA, USA) with 24-well plates. After that, placed it in an incubator at 37 ℃ with carbon dioxide for 4–6 h to form a gel. After drying the culture medium, 500 μL of serum-free medium was added to the lower chamber, and the basement membrane was hydrated for half an hour. Cell suspension was prepared by serum-free culture, and taken 100 μL into the upper chamber and add 500 μL complete culture liquid to the lower chamber. The next day, the chamber was taken out and used the cotton swab to erase the remaining cells in the upper chamber. The bottom membrane was fixed with 4% paraformaldehyde for 30 m. After 20 m dyeing with 0.1% crystal violet and cleaning with PBS, five visual fields were randomly selected under the microscope and photographed for observation and counting.
The migration assay was similar to the invasion assay, except for that Matrigel was not used in Transwell chamber.
Statistical analysis
The experimental data were analyzed by SPSS22.0 statistical analysis software. Using Student’s t-test to compare the discrepancy between two groups, and one-way ANOVA variance analysis and Tukey’s test were used to compare the mean values among three or more groups. Assessment of correlation between gene expression and clinical features was performed by chi-square test. Survival Curve was plotted by Kaplan-Meier method, and the discrepancy between groups was examined by Log-rank test. The high-expression group and the low-expression group were grouped according to the median expression level of WARS. P values <0.05 were considered as statistically significant.
Result
WARS expression was up-regulated in UM cell lines
So as to identify genes associated with metastasis in UM, we selected 63 samples from the GEO (access number: GSE22138) for analysis, including the first group named Metastasis which has 35 samples and the second group named No-Metastasis which has 28 samples. The result in ) showed that the expression of WARS in tumor metastasis group increased significantly than that in no-metastasis group (P < 0.001). This illustrated that WARS was associated with tumor metastasis and has a promoting effect in UM progression.
Figure 1. The expression level of WARS in UM. (a) Through bioinformatics analysis, we found that the expression of WARS in metastasis group was significantly higher than that in no-metastasis group (P < 0.001). (b, c, and d) The mRNA and protein expression level of WARS was up-regulated in UM cell lines (OCM-1A, MUM-2C, C918, and MUM-2B) than that in normal control cells D78 (**P < 0.01,##P < 0.01, #P < 0.05).
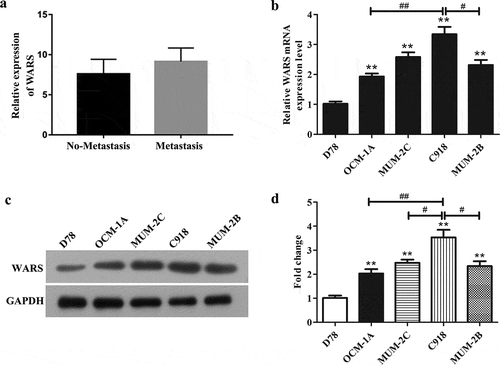
In order to further detect the mRNA and protein expression of WARS in UM cells, we conducted qRT-PCR and western blot assays. It can be seen from and ) that the mRNA and protein expression levels of WARS were markedly enhanced in human UM cell lines (OCM-1A, MUM-2C, C918, and MUM-2B) than that in normal control cells D78 (P < 0.01). Interestingly, we can see obviously that WARS was expressed differently in these cell lines OCM-1A, MUM-2C, C918, and MUM-2B. We chose C918 cells which presented the highest expression of WARS for knockdown assays and OCM-1A cells which possess a relatively lowest expression of WARS for overexpression assays.
High expression of WARS in patients with UM was associated with histological type, distant metastasis, recurrence and poor prognosis.
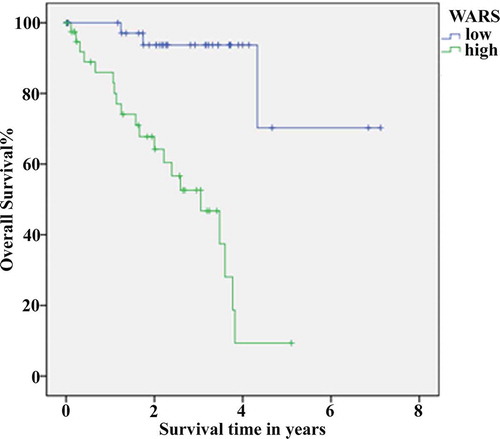
To analyze the relationship between WARS gene expression and clinical characteristics, we downloaded the RNA-Seq expression data of malignant UM from TCGA database (https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga). From the statistical analysis method of chi-square test, the expression of WARS was correlated with histological type, distant metastasis, recurrence, and death in patients with UM (, P < 0.001). After that, for the sake of investigating the relationship between WARS and prognosis, we performed survival analysis using the Kaplan-Meier method. The results in demonstrated a significant difference in overall survival of patients with UM between the high-expression and low-expression groups and revealed that high expression was associated with poor prognosis (P < 0.001). These results suggested that regulating the expression of WARS may be a new approach to the clinical treatment of UM.
Table 1. The correlation between WARS expression and clinical features of patients with uveal melanoma.
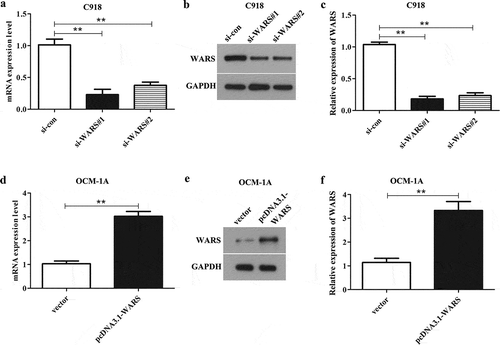
The efficiency of knockdown of WARS in C918 cells and overexpression of WARS in OCM-1A cells
To study the function of WARS, C918 cells were separately transfected with si-WARS#1 and si-WARS#2 and OCM-1A cells were transfected with pcDNA3.1-WARS. After transfection for 24 h, we performed qRT-PCR and western blotting assay to test the expression of WARS in transfected cells. The result illustrated that the mRNA and protein expression level of WARS in C918 cells transfected with si-WARS#1 or si-WARS#2 was significantly decreased than that in C918 cells transfected with si-con (), P < 0.01), and we can clearly see that the knockdown efficiency of si-WARS#1 was higher than that of si-WARS#2, so we chose si-WARS#1 for the knockdown experiments. At the same time, it can be seen from ) that transfection of pcDNA3.1-WARS in OCM-1A cells can markedly increase the mRNA and protein levels of WARS (P < 0.01).
Figure 3. The efficiency of WARS knockdown/overexpression in UM cell lines C918 and OCM-1A. (a, b, and c) The expression of WARS was markedly decreased in C918 cells after transfected with si-WARS#1 and si-WARS#2 than that in si-con group. (**P < 0.01) (d, e, and f) The expression of WARS was markedly increased in OCM-1A cells after transfected with pcDNA3.1-WARS than that in the vector group. (**P < 0.01).
Effects of altering WARS expression on cell proliferation in UM cell lines and normal D78 cell
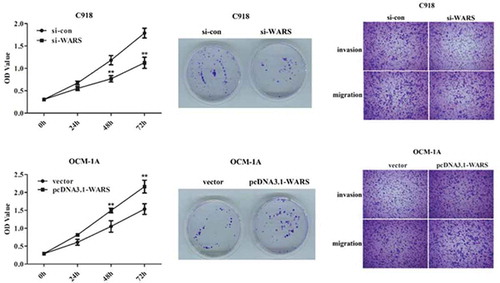
To detect the affects of knockdown or overexpression of WARS on cell proliferation, we carried out CCK8 and colony formation assay. As can be seen from the results of the CCK8 experiment, the viability was declined in C918 cells transfected with si-WARS than that transfected with si-con (), P < 0.01), whereas overexpression of WARS in OCM-1A cells can promote the viability (), P < 0.01). Subsequently, to further understand the WARS function, we examined the effect of overexpression/knockdown of WARS on the viability in normal D78 cells. The result is shown in ) that the up-regulation of WARS can increase the viability while down-regulation of WARS can decrease the viability in D78 cells (P < 0.05).
Figure 4. Deletion of WARS in C918 cell suppressed proliferation and overexpression of WARS in OCM-1A cells promoted proliferation. (a) CCK8 assay revealed that the down-regulation of WARS can reduce the cell viability in C918 cells (**P < 0.01). (b) The cell viability was obviously elevated in OCM-1A cells after overexpression of WARS (**P < 0.01). (c) Up-regulation of WARS can increase the viability while down-regulation of WARS can decrease the viability in D78 cells (P < 0.05) (d and e) Colony formation assay revealed that knockdown of WARS can reduce the ability of cells to form colonies (**P < 0.01). (f and g) Up-regulation of WARS can increase the number of colonies formed significantly than that in vector group (**P < 0.01).
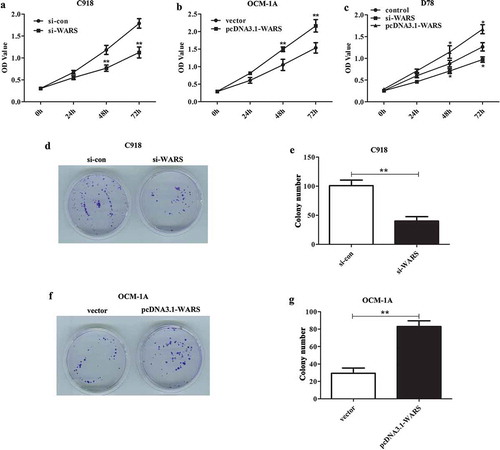
Similar results can be seen from the colony formation experiment, knockdown of WARS can inhibit the capability to form colonies in C918 cells (,, P < 0.01), and overexpression of WARS showed the opposite result (, P < 0.01). Taken together, these results demonstrate that the down-regulation of WARS can inhibit cell proliferation and thus possibly suppress the growth of UM tumors.
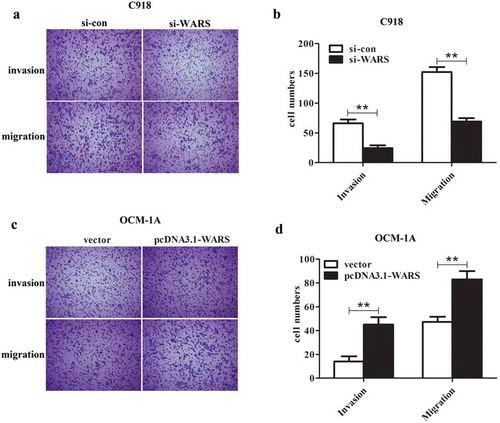
Effects of altering WARS expression on cell invasion and migration in UM
In order to explore whether knockdown or over-expression of WARS has an effect on cell migration and invasion, we carried out a Transwell assay. As shown in ), the migration and invasion of C918 cells transfected with si-WARS decreased significantly compared with the si-con group (P < 0.01). Meanwhile, we can see from ) that the upregulation of WARS can induce cell migration and invasion in OCM-1A cells. The results in this section indicated that the migration and invasion of UM cells can be regulated by regulating the expression level of WARS.
Figure 5. Deletion of WARS suppressed invasion and migration of C918 cells and overexpression of WARS promoted invasion and migration of OCM-1A cells. (a and b) Through the Transwell assay, we found that knockdown of WARS showed a significant inhibition on invasion and migration in C918 cells (**P < 0.01). (c and d) Upregulation of WARS showed a significant promotion on invasion and migration of OCM-1A cells (**P < 0.01).
PI3K/AKT/mTOR pathway mediates the regulation of WARS on the function of UM cells
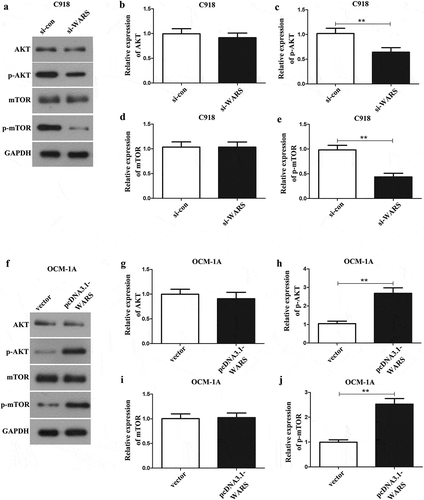
Furthermore, to confirm whether WARS plays its biological function through the PI3K/AKT/mTOR pathway, we carried out a western blot assay to measure the protein expression level of pathway-related factors. The results, as shown in and ), indicated that the phosphorylation levels of AKT and mTOR in si-WARS group showed a significant decrease than those in si-con group in C918 cells (p < 0.01), but the expression of total AKT and mTOR was not changed in C918 cells ( ( and )). Concurrently, we found that overexpression of WARS plays an opposite role in OCM-1A cells ()). Overall, these results illustrated that silencing WARS can inhibit the activation of the AKT/mTOR pathway, thereby regulating the cell proliferation, migration, and invasion of UM tumors.
Discussion
Currently, more than 80% of patients with UM will have liver metastasis and lead to death, with few effective treatment options available [Citation23]. Therefore, finding an appropriate and reasonable therapeutic target is an urgent problem to be solved. In this study, we first found from bioinformatics analysis that the expression of WARS in metastatic cells of UM patients was significantly elevated, which was related to clinical symptoms and prognosis. Subsequently, we uncovered that WARS is overexpressed in UM and promotes proliferation, migration, and invasion of UM cells partially through the PI3K/AKT signaling pathway.
WARS is abnormally expressed in various cancers and participates in biological processes such as the development of cancer. Previous studies have found that WARS was highly expressed in oral cancer [Citation14], ovarian cancer [Citation11], gastric cancers [Citation10] and cervical cancer [Citation12] compared with normal tissues, and overexpression of WARS in oral cancer improved the invasiveness of cells. Further research discovered that WARS in metastatic tumor cells shows a higher level compared with that in primary tumor cells [Citation14]. On the contrary, low expression of WARS promotes lymph node metastasis in colorectal cancer [Citation13]. In pancreatic cancer, hypoxia can down-regulate the expression of WARS and significantly increase the metastasis ability of cancer cells [Citation9]. WARS has also been reported to transmit cellular signals during human angiogenesis or protein synthesis [Citation24,Citation25]. Our study confirmed a significant increase in the expression of WARS in UM, and deletion of WARS in C918 cell suppressed proliferation, migration, and invasion, whereas overexpression of WARS in OCM-1A cells promoted these biological functions, suggesting that WARS may be a specific biomarker for clinical treatment of UM.
PI3K/AKT signaling pathway is frequently activated in cancer cells [Citation26], and the regulatory roles of PI3K/AKT signaling pathway in tumor proliferation, growth, and differentiation have been widely reported [Citation27–Citation30]. Studies have shown that AKT plays a key role in PI3K/AKT signaling pathway, and activation of PI3K can activate AKT and catalyze its phosphorylation [Citation31]. AKT phosphorylation can regulate the substrate protein of protein kinase C (PKC) and then regulate a series of physiological processes in cells [Citation31], which can be considered to be a hallmark of activation of PI3K/AKT signaling pathway [Citation32]. Recently, several reports further confirmed that activation of PI3K/AKT signaling pathway can promote the migration, proliferation of UM cancer cells [Citation21,Citation22]. Regulation of PI3K/AKT signaling pathway may be one of the effective ways to inhibit the development and metastasis of UM. Furthermore,,mTOR can regulate p70S6K which activated in UM cells, so it is another important target of PI3K [Citation22]. Moreover, E. Tzima et al. found in 2003 that T2-WARS, a biological fragment of WARS, exerts a vascular stabilizing factor by regulating the activation pathway of AKT [Citation33]. Our study demonstrates that over-expression of WARS in UM cells in vitro can promote the phosphorylation of AKT and mTOR, activate PI3K/AKT signaling pathway, and thus promote the proliferation of UM cells. Knockdown of WARS showed the opposite results. These outcomes suggested that the role of WARS in promoting malignant progression of UM cancer cells may be regulated by PI3K/AKT signaling pathway.
In conclusion, we confirmed that the expression of WARS was upregulated in UM cells and was associated with poor prognosis of UM patients. It may promote the growth of UM cells partially by activating PI3K/AKT signaling pathway, thus accelerating the development of tumors. This may provide a new strategy for the therapy of UM, but whether WARS also acts on other signaling pathways to promote the development of UM needs further study.
Authors’ contributions
YPP, YXH, and ZJ performed the experiments. YPP and YXH analyzed the data. YPP and YXH wrote the manuscript. YPP, YXH, and ZJ reviewed and edited the manuscript. All authors read and approved the final manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Aronow ME, Topham AK, Singh AD. Uveal melanoma: 5-year update on Incidence, Treatment, and Survival (SEER 1973–2013). Ocul Oncol Pathol. 2018;4:145–151.
- Posch C, Latorre A, Crosby MB, et al. Detection of GNAQ mutations and reduction of cell viability in uveal melanoma cells with functionalized gold nanoparticles. Biomed Microdevices. 2015;17:15.
- Amaro A, Gangemi R, Piaggio F, et al. The biology of uveal melanoma. Cancer Metastasis Rev. 2017;36:109–140.
- Nezu N, Goto H, Umazume K, et al. Clinical analysis of uveal melanoma. Nippon Ganka Gakkai Zasshi. 2017;121:413–418.
- Moschos MM, Dettoraki M, Androudi S, et al. The role of histone deacetylase inhibitors in uveal melanoma: current evidence. Anticancer Res. 2018;38:3817–3824.
- Stalhammar G, See TRO, Phillips S, et al. Digital image analysis of BAP-1 accurately predicts uveal melanoma metastasis. Transl Vis Sci Technol. 2019;8:11.
- Carvajal RD, Schwartz GK, Tezel T, et al. Metastatic disease from uveal melanoma: treatment options and future prospects. Br J Ophthalmol. 2017;101:38–44.
- Lee HC, Lee ES, Uddin MB, et al. Released tryptophanyl-tRNA synthetase stimulates innate immune responses against viral infection. J Virol. 2019;93.
- Paley EL, Paley DE, Merkulova-Rainon T, et al. Hypoxia signature of splice forms of tryptophanyl-tRNA synthetase marks pancreatic cancer cells with distinct metastatic abilities. Pancreas. 2011;40:1043–1056.
- Lu S, Wang LJ, Lombardo K, et al. Expression of indoleamine 2, 3-dioxygenase 1 (IDO1) and tryptophanyl-tRNA synthetase (WARS) in gastric cancer molecular subtypes. Appl Immunohistochem Mol Morphol. 2019;1.
- Morita A, Miyagi E, Yasumitsu H, et al. Proteomic search for potential diagnostic markers and therapeutic targets for ovarian clear cell adenocarcinoma. Proteomics. 2006;6:5880–5890.
- Arnouk H, Merkley MA, Podolsky RH, et al. Characterization of molecular markers indicative of cervical cancer progression. Proteomics Clin Appl. 2009;3:516–527.
- Ghanipour A, Jirstrom K, Ponten F, et al. The prognostic significance of tryptophanyl-tRNA synthetase in colorectal cancer. Cancer Epidemiol Biomarkers Prev. 2009;18:2949–2956.
- Lee CW, Chang KP, Chen YY, et al. Overexpressed tryptophanyl-tRNA synthetase, an angiostatic protein, enhances oral cancer cell invasiveness. Oncotarget. 2015;6:21979–21992.
- Seshaiah P, Andrew DJ, Kimble J. WRS-85D: A tryptophanyl-tRNA synthetase expressed to high levels in the developing Drosophila salivary gland. Mol Biol Cell. 1999;10:1595–1608.
- Yang D, Nakada-Tsukui K, Ohtani M, et al. Identification and cloning of genes associated with the guinea pig skin delayed-type hypersensitivity reaction. J Biochem. 2001;129:561–568.
- Wakasugi K, Slike BM, Hood J, et al. A human aminoacyl-tRNA synthetase as a regulator of angiogenesis. Proc Natl Acad Sci U S A. 2002;99:173–177.
- Stana A, Vodnar DC, Marc G, et al. Antioxidant activity and antibacterial evaluation of new thiazolin-4-one derivatives as potential tryptophanyl-tRNA synthetase inhibitors. J Enzyme Inhib Med Chem. 2019;34:898–908.
- Jin M. Unique roles of tryptophanyl-tRNA synthetase in immune control and its therapeutic implications. Exp Mol Med. 2019;51:1.
- Li Y, Sun D, Sun W, et al. Ras-PI3K-AKT signaling promotes the occurrence and development of uveal melanoma by downregulating H3K56ac expression. J Cell Physiol. 2019.
- Ye M, Hu D, Tu L, et al. Involvement of PI3K/Akt signaling pathway in hepatocyte growth factor-induced migration of uveal melanoma cells. Invest Ophthalmol Vis Sci. 2008;49:497–504.
- Babchia N, Calipel A, Mouriaux F, et al. The PI3K/Akt and mTOR/P70S6K signaling pathways in human uveal melanoma cells: interaction with B-Raf/ERK. Invest Ophthalmol Vis Sci. 2010;51:421–429.
- Chattopadhyay C, Kim DW, Gombos DS, et al. Uveal melanoma: from diagnosis to treatment and the science in between. Cancer. 2016;122:2299–2312.
- Yang XL, Schimmel P, Ewalt KL. Relationship of two human tRNA synthetases used in cell signaling. Trends Biochem Sci. 2004;29:250–256.
- Sajish M, Zhou Q, Kishi S, et al. Trp-tRNA synthetase bridges DNA-PKcs to PARP-1 to link IFN-gamma and p53 signaling. Nat Chem Biol. 2012;8:547–554.
- Spangle JM, Roberts TM, Zhao JJ. The emerging role of PI3K/AKT-mediated epigenetic regulation in cancer. Biochim Biophys Acta Rev Cancer. 2017;1868:123–131.
- Yang SX, Polley E, Lipkowitz S. New insights on PI3K/AKT pathway alterations and clinical outcomes in breast cancer. Cancer Treat Rev. 2016;45:87–96.
- Ham J, Lim W, Kim K, et al. Gentisyl alcohol inhibits proliferation and induces apoptosis via mitochondrial dysfunction and regulation of MAPK and PI3K/AKT pathways in epithelial ovarian cancer cells. Mar Drugs. 2019;17.
- Yang H, Liu JX, Shang HX, et al. Qingjie Fuzheng granules inhibit colorectal cancer cell growth by the PI3K/AKT and ERK pathways. World J Gastrointest Oncol. 2019;11:377–392.
- Li B, Yang J, Lu Z, et al. A study on the mechanism of rapamycin mediating the sensitivity of pancreatic cancer cells to cisplatin through PI3K/AKT/mTOR signaling pathway. J Buon. 2019;24:739–745.
- Wang T, Seah S, Loh X, et al. Simvastatin-induced breast cancer cell death and deactivation of PI3K/Akt and MAPK/ERK signalling are reversed by metabolic products of the mevalonate pathway. Oncotarget. 2016;7:2532–2544.
- Xu Q, Simpson SE, Scialla TJ, et al. Survival of acute myeloid leukemia cells requires PI3 kinase activation. Blood. 2003;102:972–980.
- Tzima E, Reader JS, Irani-Tehrani M, et al. Biologically active fragment of a human tRNA synthetase inhibits fluid shear stress-activated responses of endothelial cells. Proc Natl Acad Sci U S A. 2003;100:14903–14907.