ABSTRACT
Zinc finger protein 521 (Zfp521) is a key transcriptional factor in regulation of hematopoiesis. SUMOylation, a protein post-translational modification process, plays important roles in various biological process including hematopoiesis. However, whether Zfp521 can be SUMOylated and how it affects hematopoiesis is unknown. In this study, we confirmed that Zfp521 can be modified by SUMO1 and lysine 1146 was the primary SUMOylation site. Under homeostatic condition, Zfp521 SUMOylation-deficient mice had normal mature blood cells and primitive cells. However, in bone marrow (BM) transplantation assay, recipient mice transplanted with BM cells from Zfp521 SUMOylation-deficient mice had a significantly decreased R2 population of erythroid lineage in BM and spleen compared with those transplanted with BM cells from wild-type mice. Our results found a novel function of Zfp521 SUMOylation in erythroid reconstitution under stress, which might be a new therapeutic target in future.
Graphical abstract
Zfp521 was modified by SUMO1 on lysine 1146
Hematopoiesis develops in the embryonic stage, produces and replenishes the whole blood cellular component throughout adulthood in a well-characterized and stepwise way [Citation1]. In adult mammalian, hematopoiesis occurs in the bone marrow (BM) from hematopoietic stem cells (HSCs) which are capable of indefinite self-renewal and differentiation to generate multipotent progenitor cells (MPPs). MPPs then commit to myeloid, lymphoid, or erythroid/megakaryocytic lineage and finally differentiate into mature blood cells after different hierarchical stages [Citation2,Citation3]. The process of hematopoiesis is tightly regulated by intrinsic molecular mechanisms and extrinsic signals from the microenvironment [Citation4–Citation9]. In clinical condition such as bone marrow transplantation (BMT), the hematopoiesis in BM needs to be reconstituted to produce enough mature blood cells again for the body [Citation10–Citation12]. The regulation of hematopoietic reconstitution is being increasingly emphasized with extensive usage of BMT for treating leukemia or other cancer [Citation10,Citation13–Citation16].
SUMOylation is a conserved dynamic protein post-translational modification process. SUMO (small ubiquitin-like modifier) conjugation is carried out by a multistep enzymatic cascade reaction consisting of a SUMO-activating enzyme (E1), Aos1/Uba2, a unique SUMO-conjugating enzyme (E2), Ubc9, and three families of SUMO ligases (E3), which eventually attach SUMO to the substrates [Citation17–Citation19]. De-SUMOylation process is mediated by SENP (SUMO-specific protease) family members, including SENP1, 2, 3, 5, 6 and 7 [Citation20,Citation21]. Protein SUMOylation plays important roles in different biological and pathological process by affecting many aspects of protein functions, including transcription regulation, genome integrity, and subcellular localization [Citation18]. Previous studies have demonstrated that SUMOylation of HIF-1a, STAT5, c-Myb, etc. participated in the regulation of hematopoiesis [Citation22–Citation24].
Zinc finger protein 521 (Zfp521 in mice or ZNF521 in humans; also known as EVI3 and EHZF) is a transcriptional regulator containing an N terminal transcriptional repressor motif and C2H2-type zinc finger which abundantly expresses in immature cell populations, such as hematopoietic progenitors [Citation25]. Previous study demonstrated that Zfp521 participates in the regulation of hematopoiesis [Citation26–Citation29]. It modulates erythroid cell differentiation through directly binding with GATA-1 [Citation27]. Zfp521 can also regulate B-cell development by binding to EBF1 via its carboxyl terminal portion, and regulate B cell viability via cyclin D1 gene expression [Citation28,Citation29].
Whether Zfp521 can be SUMOylated and how this post-translational modification of Zfp521 participates in the regulation of hematopoiesis is an important question which needs to be investigated. Here, we demonstrated that Zfp521 can be SUMOylated. Although the deficient of SUMOylation of Zfp521 did not affect hematopoiesis in homeostatic condition, it can impair erythroid lineage reconstitution under stress.
Materials and methods
Plasmids and antibodies
Plasmid FLAG-Zfp521 was purchased from Bio-Atom (Chengdu, China). Plasmids HA-SUMO1 was generated as previously described [Citation22]. The potential SUMOylation residue of lysine 1146 (Lys-1146) was replaced with arginine (K1146R) using the QuichChangeTM site-directed mutagenesis kit (Stratagene, La Jolla, CA, USA). The primers used to construct plasmid FLAG-Zfp521-K1146R were: forward, 5ʹ-AGCTGCAACGTTAGGTTTGAGTCTGAA-3ʹ; reverse, 5ʹ-TTCAGACTCAAACCTAACGTTGCAGCT-3ʹ. All the plasmids were confirmed by DNA sequencing. Anti-FLAG and anti-HA antibodies were purchased from Sigma (St. Louis, MO, USA); anti-SUMO1 and anti-Zfp521 antibodies were from Abcam (Cambridge, MA, USA); and anti-β-actin antibody was from Proteintech Group (Wuhan, China).
Cell culture and transfection
HEK293T cell lines were purchased from the Chinese Academy of Science cell bank (Shanghai, China). The cells were cultured in DMEM medium (Hyclone), supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin (Gibco) at 37°C in 5% CO2 incubator. The desired plasmids were transfected into HEK293T cells with Lipofectamine 2000 (Invitrogen) following the protocol provided by the manufacturer.
Immunoprecipitation and immunoblotting
Cells were lysed at 36 h after transfection in radioimmune precipitation assay (RIPA) buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1% NP-40, 0.1% SDS, 1 mM EDTA, 10 mM N-ethylmaleimide, 1 mM PMSF and a cocktail of protease inhibitors) on ice for 30 min. The cell lysates were sonicated at 25% AMPL for 10s three times and centrifugated at 14,000 g for 10 min. The supernatants were incubated with 10 µL anti-FLAG M2 agarose beads or 10 µL anti-HA agarose beads (Sigma) at 4°C for 4 h on a rotating shaker. After washing beads with the above-mentioned RIPA buffer, beads were mixed with 2 × SDS and boiled at 100°C for 10 min. The immunoprecipitated samples and total extracts were resolved by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and blotted on a PVDF membrane (Millipore). The proteins of interest were detected with corresponding antibodies according to manufacturer’s instructions and immunoblots were analyzed using the LAS-4000 system (Fujifilm) [Citation30].
Generation of Zfp521 SUMOylation-deficiency mice
C57BL/6 mice model with Zfp521 SUMOylation-deficiency was generated by changing the Lys-1146 to Arg-1146 (K1146R) in the endogenous Zfp521 mice locus using CRISPR/Cas-mediated genomic engineering (Cyagen Biosciences Inc). To introduce K1146R mutation, guide RNA (gRNA) targeting the coding region of K1146 and donor oligo consisted of targeting sequence and 120 bp homologous arms flanking the cutting sites were designed. The K1146R mutation sequence (AAG to AGG) was introduced in the donor oligo. To prevent re-cutting of the locus by sgRNA, a silent mutation (ATC to ATA) was also introduced to gRNA sequence in the donor oligo. Cas9 mRNA, gRNA and donor oligo were co-injected into zygotes for KI mouse production. The resulting heterozygous mutant females were mated with wild-type (WT) C57BL/6N male to get the next generation (F1) heterozygous mutant mouse. The genotype was screened by PCR and DNA sequencing analysis. The sequences for gRNA, homologous arms, PCR genotyping and DNA sequencing are shown in . The Zfp521 homozygous mutant mice (referred to as “Zfp521mutant mice”) was used as experimental mice, and age- and gender-matched WT C57BL/6N mice (referred to as “Zfp521WT mice”) was used as control. All mouse experiments were carried out in accordance with the guidelines for the Care and Use of Laboratory Animals and approved by the Animal Ethics Committee of the First Affiliated Hospital of Army Medical University.
Table 1. Sequences of guide RNA, donor oligo, PCR genotyping primer and DNA sequencing primer.
RT-qPCR
Zfp521 mRNA expression levels in peripheral blood and BM of Zfp521mutant mice and Zfp521WT mice were detected with RT-qPCR. Total RNA was extracted from peripheral blood and BM cells by Trizol kit (Invitrogen) and treated with DNase (Promega). 2 µg RNA was reverse-transcribed to synthesis complementary DNA (cDNA) with the cDNA synthesis kit (Takara). Quantitative PCR (qPCR) was performed using the CFX96 Real-Time PCR Detection System (Bio-Rad Laboratories). β-actin was used as an endogenous control to normalize Zfp521 expression.
Complete blood counts
Peripheral blood of Zfp521mutant mice and Zfp521WT mice were collected by tail bleeding into tubes containing EDTA. Complete blood counts and differentials were detected by hematology analyzer Hemavet 950FS (Drew Scientific, USA) according to manufacturer’s instructions.
Flow cytometry analysis
Single-cell suspensions were prepared from BM, thymus, and spleen. After lysis of red blood cells (RBC) by ammonium chloride-potassium buffer (Solarbio, Beijing, China), remaining cells were incubated with antibodies on ice for 20 min. The following antibodies conjugated to fluorochrome or biotin against mouse molecules were used: Gr-1, Ter119, B220, CD19, Rat-IgM, IL-7R and CD3 for lineage markers; Streptavidin-PE-Cy5, PE-Sca-1, APC-Cy7-c-Kit, PE-Cy7-CD48 and APC-CD150 for HSC, hematopoietic progenitor cell (HPC) and LSK analysis; Streaptavidin-APC-Cy7, PE-Sca-1, APC-c-Kit, PE-Cy7-CD16/32 and eFluor450-CD34 for common myeloid progenitor (CMP), granulocyte-macrophage progenitor (GMP) and megakaryocyte-erythroid progenitor (MEP) analysis; Streptavidin-APC-Cy7, PE-Sca-1, APC-c-Kit and PE-Cy5-Flt3 for lymphoid-primed multipotent progenitor pool cell (LMPP) analysis; PE-Gr-1 and APC-Mac-1 for myeloid cell analysis; APC-Ter119 and PE-CD71 for erythroid cell analysis; PE-B220 and APC-IgM for B cell analysis; PE-CD4 and APC- CD8 for mature T cell analysis. All of the antibodies were purchased from eBioscience, except APC-CD150 from Biolegend. Flow cytometry analysis was performed on BD FACS Calibur and all data were analyzed by FlowJo10 software (Tree Star, Inc.).
Bone marrow transplantation
C57BL/6NCR (Ly5.1) recipients (6 to 8 weeks old, about 20 g) were purchased from Army medical university. Prior to transplantation, the recipients were lethally irradiated with a total dose of 13 Gy, which was split into two doses of 6.5 Gy given every 3 h. Total 2 × 106 freshly harvested BM cells from Zfp521mutant mice or Zfp521WT mice, which have been depleted of RBC through resuspending in hypotonic buffer (0.16 MNH 4Cl, 0.17 M Tris-HCl [pH 7.4]), were injected into the retro-orbital venous sinus of lethally irradiated Ly5.1 recipients. BM, thymus and spleen cells were obtained from recipients at 2 months after transplantation and analyzed by flow cytometric analysis.
Statistical analysis
Data are expressed as mean ± standard deviation (SD). Statistical significance of differences between groups was determined using two-sided Student’s t-test. p < 0.05 was considered as statistically significant. All statistical analyses were performed using SPSS 20.0 software (Chicago, IL, USA).
Results
Zfp521 was modified by SUMO1 on lysine residue 1146
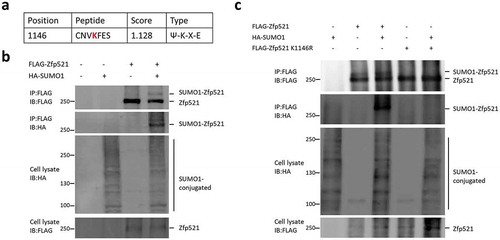
Through using SUMOylation site prediction software of SUMOsp 2.0 [Citation31], we found there was one SUMO consensus sequence (Ψ-K-X-E) within Zfp521 ()), suggesting this protein may be SUMOylated. To test whether Zfp521 is indeed SUMOylated, HEK293T cells were transiently co-transfected with plasmids of FLAG-Zfp521 or empty vector (pTango) and plasmids of HA-SUMO1 or empty vector (pCDNA3.1). After culture for 36 h, FLAG-Zfp521 was immunoprecipitated with anti-FLAG M2 agarose beads and then immunoblotting with anti-HA or anti-FLAG antibodies. As shown in ), SUMOylated Zfp521 band was detected in cells transfected with FLAG-Zfp521 and HA-SUMO1, but not in cells transfected with either FLAG-Zfp521 or HA-SUMO1 alone, suggesting Zfp521 can be modified by SUMO1. As noted in ), Lys-1146 may be a primary SUMO1 conjugation amino acid residue. To confirm this, we mutated the conserved lysine residue to arginine residue and detected SUMOylation status of the Zfp521-K1146R mutant. As shown in ), SUMOylated Zfp521 band was clearly observed in cells co-transfected with HA-SUMO1 and wild-type Zfp521. However, the SUMOylated band was completely abolished when cells transfected with HA-SUMO1 and the Zfp521-K1146R mutant. This result demonstrates Lys-1146 is the primary SUMOylation site of Zfp521.
Figure 1. Zfp521 was modified by SUMO1 on lysine 1146. (a) Potential SUMO modification site in Zfp521 was predicted by SUMOsp 2.0 software. (b) Zfp521 SUMOylation status was measured by immunoprecipitation (IP) and immunoblotting (IB) after co-transfection of HEK293T cells with HA-SUMO1 and FLAG-Zfp521. (c) Zfp521 SUMOylation status was detected by IP and IB after co-transfection of HEK293T cells with HA-SUMO1, FLAG-Zfp521 or Zfp521-K1146R mutant.
Generation of Zfp521 SUMOylation-deficient mice
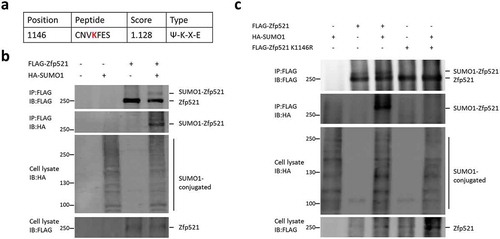
To investigate the in vivo function of Zfp521 SUMOylation in normal hematopoiesis, we generated Zfp521 SUMOylation-deficient mice by CRISPR/Cas-mediated genomic engineering. As was demonstrated above, Zfp521 can be SUMOylated at Lys-1146. The lysine residue is located in perfect SUMOylation consensus sequence on exon 4 of Zfp521 gene in mice chromosome 18. To abolish Zfp521 SUMOylation in mice, the lysine was mutated to arginine to generate heterozygous and homozygous K1146 mutant mice (). The genotype of the mice was determined by PCR () and further confirmed by sequencing () of genomic DNA isolated from mice tail cells. RT-qPCR and immunoblotting showed that the mutant Zfp521 was expressed at a level comparable with that of wild-type Zfp521 in the PB and BM (,).
Figure 2. Zfp521 SUMOylation-deficient mice were generated. (a) Schematic representation of the gene structure of Zfp521. The primary SUMOylation site of lysine 1146 was mutated to arginine (K1146R). (b) The genotypes of mice were determined by PCR. (c) The genotypes of Zfp521 wild-type (Zfp521WT), K1146R heterozygote mutant (Zfp521±) and homozygous mutant (Zfp521mutant) mice were confirmed by DNA Sequence. (d) Zfp521 mRNA levels were detected by RT-qPCR in peripheral blood (PB) and bone marrow (BM) of Zfp521WT and Zfp521mutant mice at 7 weeks of age, expressed as percent of expression in Zfp521WT mice (n = 3 per group). (e) Zfp521 protein levels were detected by western blotting in PB and BM of Zfp521mutant and Zfp521WT mice at 7 weeks of age.
Zfp521 SUMOylation-deficient mice had normal mature blood cells
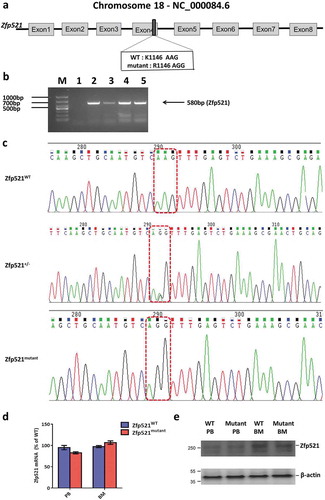
After the mice with Zfp521 SUMOylation site mutation (K1146R) were constructed, we firstly investigated the effect of Zfp521 SUMOylation on mature blood cells. Complete blood counts of Zfp521mutant mice and Zfp521WT mice at 7 weeks of age were detected by Hemavet 950FS blood routine analyzer. Absolute number of white blood cells (WBC), neutrophils (NE), lymphocytes (LY), monocytes (MO), eosinophils (EO), basophils (BA), platelets (PLT), RBC and hemoglobin (HB) in peripheral blood were comparable between Zfp521mutant mice and Zfp521WT mice (,). Erythrocyte, myeloid cells, B cells and T cells in BM, spleen or thymus of Zfp521mutant mice and Zfp521WT mice were detected by flow cytometry analysis. It was found that there were also no statistically significant differences in absolute number of these mature blood cells between Zfp521mutant mice and Zfp521WT mice (–). These results suggest that Zfp521 SUMOylation deficient did not affect mature blood cells number under homeostatic condition.
Figure 3. Zfp521 SUMOylation-deficient mice had normal peripheral blood counts. (a and b) Absolute number of white blood cells (WBC), neutrophils (NE), lymphocytes (LY), monocytes (MO), eosinophils (EO), basophils (BA) (a), platelets (PLT), red blood cells (RBC), and hemoglobin (HB) (b) in peripheral blood of Zfp521mutant and Zfp521WT mice at 7 weeks of age. Data were from two independent experiments (n = 4, per experiment) and present as mean ± SD.
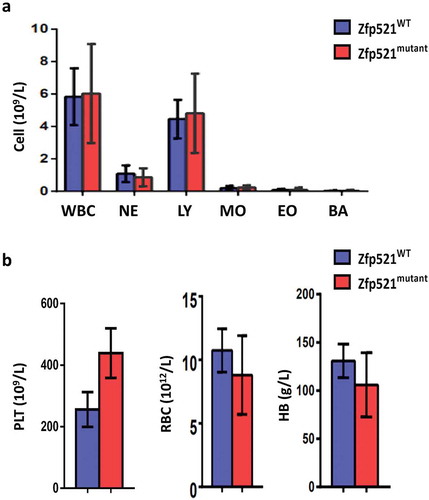
Figure 4. Zfp521 SUMOylation-deficient mice had normal mature blood cells in bone marrow, spleen and thymus. (a-c) Flow cytometric analysis and absolute number of different development stages of erythrocytes (a), myeloid cells (b), and B cells (c) in bone marrow (BM) and spleen of Zfp521mutant and Zfp521WT mice at 7 weeks of age. (d) Flow cytometric analysis and absolute numbers of T cells in BM, spleen, and thymus of Zfp521mutant and Zfp521WT mice at 7 weeks of age. Data were from two independent experiments (n = 4, per experiment) and present as mean ± SD.
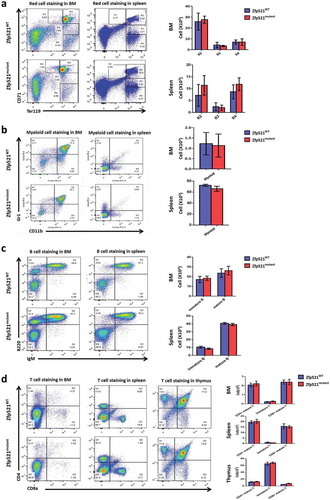
Zfp521 SUMOylation-deficient mice had normal primitive cells in BM
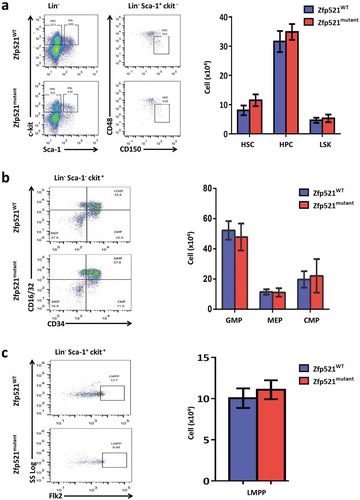
We further investigated the effect of Zfp521 SUMOylation on the production of primitive cells in BM. Hematopoietic stem cells (HSC), hematopoietic progenitor cells (HPC), LSK cells, common myeloid progenitors (CMP), more committed myeloid progenitor cells including granulocyte-macrophage progenitors (GMP), megakaryocyte-erythroid progenitors (MEP), and lymphoid-primed multipotent progenitor pool cells (LMPP) were detected in BM of Zfp521mutant mice and Zfp521WT mice by flow cytometry analysis. The results showed that there were no statistically significant differences in absolute number of these primitive cells in BM between Zfp521mutant mice and Zfp521WT mice (), suggesting Zfp521 SUMOylation deficient did not affect hematopoietic differentiation in BM under homeostatic condition.
Figure 5. Zfp521 SUMOylation-deficient mice had normal primitive cells in bone marrow. (a-c) Flow cytometry analysis and absolute number of hematopoietic stem cells (HSC), hematopoietic progenitor cells (HPC) and LSK cells (a), and granulocyte-macrophage progenitors (GMP), megakaryocyte-erythroid progenitors (MEP) and common myeloid progenitors (CMP) (b), and lympho-primed pluripotent progenitor cells (LMPP) in BM of Zfp521mutant and Zfp521WT mice at 7 weeks of age. Data were from two independent experiments (n = 4, per experiment) and present as mean ± SD.
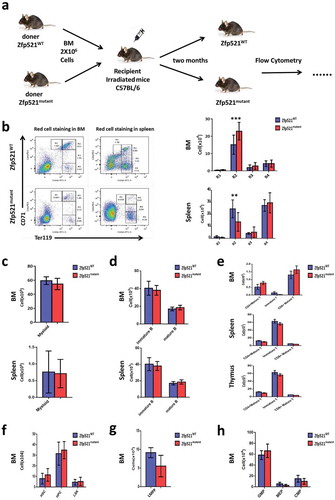
Zfp521 SUMOylation is important for erythroid hematopoietic recovery under stress
To test whether SUMOylation of Zfp521 participates in the regulation of hematopoietic reconstitution under stress, we performed BMT assay through injecting equal numbers of BM cells from Zfp521mutant mice and Zfp521WT mice to lethally irradiated recipients, and detected hematopoietic stem cells and mature blood cells in these mice by flow cytometric analysis after 2 months of transplantation (). The results showed that recipient mice transplanted with BM cells from Zfp521mutant mice had a statistically significantly decreased R2 population of erythroid lineage in BM and spleen compared with those transplanted with BM cells from Zfp521WT mice (), although the differences of other cells were not significant (–), suggesting that SUMOylation of Zfp521 facilitates erythroid hematopoietic reconstitution process under stress.
Figure 6. Erythroid hematopoietic recovery of transplanted mice was affected by Zfp521 SUMOylation. (a) The diagram for the experimental design. (b) Flow cytometric analysis and absolute number of different development stages of erythrocytes in BM and spleen of 8-week old recipient mice after transplantation BM cells from Zfp521mutant mice and Zfp521WT mice. (c and d) Absolute number of myeloid cells (c) and B cells (d) in BM and spleen of 8-week old recipient mice after transplantation BM cells from Zfp521mutant mice and Zfp521WT mice. (e) Absolute number of T cells in the BM, spleen, and thymus of 8-week old recipient mice after transplantation BM cells from Zfp521mutant mice and Zfp521WT mice. (f-h) Absolute number of HPC, LSK and HSC cells (f), LMPP cells (g), and CMP, GMP and MEP cells (h) in BM of 8-week old recipient mice after transplantation BM cells from Zfp521mutant mice and Zfp521WT mice. Data were from three independent experiments (n = 3, per experiment) and present as mean ± SD. (***p < 0.001).
Discussion
Many transcription factors regulating hematopoiesis or reconstitution have been identified and mechanistically characterized [Citation32]. However, whether post-translational modification of these factors participates in the regulation of hematopoiesis or reconstitution is less understood. This study, along with other recent reports, provides a new clue for SUMOylation of transcriptional factor participates in the regulation of hematopoiesis or reconstitution.
In this study, we obtained several lines of evidences that support the conclusion that deficiency of SUMOylation of Zfp521 did not affect hematopoiesis in homeostatic condition, but can regulate hematopoietic reconstitution under stress. Firstly, we demonstrated that Zfp521 can be SUMOylated and found out that Lys-1146 on Zfp521 was the conjugation site for SUMOylation by biochemical and molecular experiments. Then, we generated the Zfp521 K1146R point mutation mice to find out that deficiency of SUMOylation of Zfp521 did not affect hematopoiesis in homeostatic condition. The number of mature cells including WBC, NE, LY, MO, EO, BA, PLT, RBC, myeloid cells, B cells and T cells in peripheral blood, thymus, and spleen did not differ between Zfp521mutant mice and Zfp521WT mice. The number of HSC, HPCs, LSK cells, CMP, GMP, MEP, and LMPP in BM also were similar between wild-type mice and mutant mice. At last, we found that deficient of Zfp521 SUMOylation caused markedly decreased R2 population of erythroid lineage in BM and spleen of transplantation recipient mice, which suggesting Zfp521 SUMOylation facilitates erythroid reconstitution under stress.
We speculated that why SUMOylation of Zfp521 participated in the regulation of hematopoietic reconstitution under stress but did not affect hematopoiesis in homeostatic condition may on the following aspects. First, without proper SUMOylation of Zfp521, the HSCs can function rightly in homeostatic condition, but the capacity of HSCs to deal with stress might decrease [Citation33]. Second, radiation before BMT may cause injury to microenvironment (niche) in BM which includes not only many cellular components but also various cytokines [Citation34], then the structural and functional interactions between HSCs and niche are being impaired. This might cause insufficient extrinsic signals to support the hematopoietic recovery in the Zfp521 K1146R mutant state. Third, the radiation can cause neuro-endocrinal change in recipient mice which is also crucial to erythroid differentiation [Citation35]. Taken together, why SUMOylation of Zfp521 participated in the regulation of erythroid reconstitution after BMT but did not affect hematopoiesis in homeostatic condition may be due to complex intrinsic factors, extrinsic factors and their interaction, the underlying mechanisms need to be further explored.
Protein SUMOylation is a dynamic process which participates in a lot of biological and pathological process, great effort has been made trying to target SUMOylation and regulating the specific biological process or even clinical situations [Citation36–Citation38]. SUMO conjugation is carried out by an enzymatic cascade reaction consisting of activating enzyme (E1), conjugating enzyme (E2), and ligases (E3). De-SUMOylation process is mediated by SENPs family members, including SENP1, 2, 3, 5, 6 and 7 [Citation17,Citation19–Citation21]. Various inhibitors or agonists have been developed to regulate this process and in order to affect the downstream biological functions. Puromycin can induce SUMO-1/2/3 accumulation and affect PML nuclear body assembly/disassembly, which is helpful for understanding nuclear sequestration of aberrant proteins and may provide clues for the elimination of toxic-aggregated protein species involved in numerous neurodegenerative diseases and other disorders [Citation39]. Adenosine can mediate SUMOylation of IкBα. The SUMOylated IкBα form is resistant to signal-induced degradation, consequently halting NFкB activation and then the cells and tissues acquires long-lasting anti-inflammatory properties [Citation40]. SUMO-mimicking peptides have been invented and the investigators found that, once the peptides were conjugated to SAE and Ubc9, they blocked full-length SUMO from entering the cascade. These peptides can thus function as mechanism-based inhibitors of the protein SUMOylation reaction [Citation41]. A highly specific covalent allosteric inhibitor (COH000) for E1 enzyme has also been developed and demonstrated its anti-tumor activity in preclinical models of colorectal cancer [Citation42,Citation43]. Multiple SENPs inhibitors have also been reported in the previous report with different intensities and different biological effects [Citation44,Citation45]. Along with these effective inhibitors or agonists, in consideration of the participation of the SUMOylation of transcriptional factors such as HIF-1a, STAT5, CEBPα, c-Myb and the newly defined Zfp521 in hematopoiesis or reconstitution, targeted manipulation of the SUMOylation status of certain protein might be useful for specific regulating hematopoiesis or reconstitution.
In summary, this study unveiled a function of SUMOylation of Zfp521 in erythroid reconstitution under stress in mice. Our study provides new insight into hematopoietic recovery, which may have targeted therapeutically implications in regeneration of normal hematopoiesis in certain circumstances.
Author contribution
J Chen, Y Hou and X Li conceived and designed the experiments. Y Zhang, S Xu, M Xie, Z Chen, Y Ma, and G Wu performed the experiments. Y Zhang, X Huang, C Luo, Z Huang, Y Sun and Y Huang analyzed the data. J Chen, Y Hou and X Li drafted the manuscript. All authors read and approved the final manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Dzierzak E, Bigas A. Blood development: hematopoietic stem cell dependence and independence. Cell Stem Cell. 2018;22(5):639–651.
- Zhang Y, Gao S, Xia J, et al. Hematopoietic hierarchy – an updated roadmap. Trends Cell Biol. 2018;28(12):976–986.
- Dharampuriya PR, Scapin G, Wong C, et al. Tracking the origin, development, and differentiation of hematopoietic stem cells. Curr Opin Cell Biol. 2017;49:108–115.
- Zon LI. Intrinsic and extrinsic control of haematopoietic stem-cell self-renewal. Nature. 2008;453(7193):306–313.
- Takizawa H, Manz MG. Impact of inflammation on early hematopoiesis and the microenvironment. Int J Hematol. 2017;106(1):27–33.
- Shimoto M, Sugiyama T, Nagasawa T. Numerous niches for hematopoietic stem cells remain empty during homeostasis. Blood. 2017;129(15):2124–2131.
- Pardo-Saganta A, Tata PR, Law BM, et al. Parent stem cells can serve as niches for their daughter cells. Nature. 2015;523(7562):597–601.
- Wei Q, Frenette PS. Niches for hematopoietic stem cells and their progeny. Immunity. 2018;48(4):632–648.
- Wu J, Zhang W, Ran Q, et al. The differentiation balance of bone marrow mesenchymal stem cells is crucial to hematopoiesis. Stem Cells Int. 2018;2018:1540148.
- Gordon MY, Blackett NM. Reconstruction of the hematopoietic system after stem cell transplantation. Cell Transplant. 1998;7(4):339–344.
- Storek J, Geddes M, Khan F, et al. Reconstitution of the immune system after hematopoietic stem cell transplantation in humans. Semin Immunopathol. 2008;30(4):425–437.
- Ogonek J, Kralj Juric M, Ghimire S, et al. Immune reconstitution after allogeneic hematopoietic stem cell transplantation. Front Immunol. 2016;7:507.
- Wang C, Zhang B, Wang S, et al. Recombinant human thrombopoietin promotes hematopoietic reconstruction after severe whole body irradiation. Sci Rep. 2015;5:12993.
- Baron F, Nagler A. Novel strategies for improving hematopoietic reconstruction after allogeneic hematopoietic stem cell transplantation or intensive chemotherapy. Expert Opin Biol Ther. 2017;17(2):163–174.
- van der Maas NG, Berghuis D, van der Burg M, et al. B cell reconstitution and influencing factors after hematopoietic stem cell transplantation in children. Front Immunol. 2019;10:782.
- de Koning C, Nierkens S, Boelens JJ. Strategies before, during, and after hematopoietic cell transplantation to improve T-cell immune reconstitution. Blood. 2016;128(23):2607–2615.
- Han ZJ, Feng YH, Gu BH, et al. The post-translational modification, SUMOylation, and cancer (Review). Int J Oncol. 2018;52(4):1081–1094.
- Yang Y, He Y, Wang X, et al. Protein SUMOylation modification and its associations with disease. Open Biol. 2017;7(10):170167.
- Wilson VG. Introduction to Sumoylation. Adv Exp Med Biol. 2017;963:1–12.
- Kunz K, Piller T, Muller S. SUMO-specific proteases and isopeptidases of the SENP family at a glance. J Cell Sci. 2018;131(6):jcs211904.
- Mukhopadhyay D, Dasso M. Modification in reverse: the SUMO proteases. Trends Biochem Sci. 2007;32(6):286–295.
- Cheng J, Kang X, Zhang S, et al. SUMO-specific protease 1 is essential for stabilization of HIF1alpha during hypoxia. Cell. 2007;131(3):584–595.
- Van Nguyen T, Angkasekwinai P, Dou H, et al. SUMO-specific protease 1 is critical for early lymphoid development through regulation of STAT5 activation. Mol Cell. 2012;45(2):210–221.
- Saether T, Pattabiraman DR, Alm-Kristiansen AH, et al. A functional SUMO-interacting motif in the transactivation domain of c-Myb regulates its myeloid transforming ability. Oncogene. 2011;30(2):212–222.
- Fleenor CJ, Arends T, Lei H, et al. Zinc finger protein 521 regulates early hematopoiesis through cell-extrinsic mechanisms in the bone marrow microenvironment. Mol Cell Biol. 2018;38(17):e00603–17.
- Mega T, Lupia M, Amodio N, et al. Zinc finger protein 521 antagonizes early B-cell factor 1 and modulates the B-lymphoid differentiation of primary hematopoietic progenitors. Cell Cycle. 2011;10(13):2129–2139.
- Matsubara E, Sakai I, Yamanouchi J, et al. The role of zinc finger protein 521/early hematopoietic zinc finger protein in erythroid cell differentiation. J Biol Chem. 2009;284(6):3480–3487.
- Kiviranta R, Yamana K, Saito H, et al. Coordinated transcriptional regulation of bone homeostasis by Ebf1 and Zfp521 in both mesenchymal and hematopoietic lineages. J Exp Med. 2013;210(5):969–985.
- Al Dallal S, Wolton K, Hentges KE. Zfp521 promotes B-cell viability and cyclin D1 gene expression in a B cell culture system. Leuk Res. 2016;46:10–17.
- Wyatt D, Malik R, Vesecky AC, et al. Small ubiquitin-like modifier modification of arrestin-3 regulates receptor trafficking. J Biol Chem. 2011;286(5):3884–3893.
- Xue Y, Zhou F, Fu C, et al. SUMOsp: a web server for sumoylation site prediction. Nucleic Acids Res. 2006;34(WebServer issue):W254–7.
- Aggarwal R, Lu J, Pompili VJ, et al. Hematopoietic stem cells: transcriptional regulation, ex vivo expansion and clinical application. Curr Mol Med. 2012;12(1):34–49.
- Ganuza M, McKinney-Freeman S. Hematopoietic stem cells under pressure. Curr Opin Hematol. 2017;24(4):314–321.
- Yu VW, Scadden DT. Hematopoietic stem cell and its bone marrow niche. Curr Top Dev Biol. 2016;118:21–44.
- Stone HB, Coleman CN, Anscher MS, et al. Effects of radiation on normal tissue: consequences and mechanisms. Lancet Oncol. 2003;4(9):529–536.
- Bialik P, Wozniak K. SUMO proteases as potential targets for cancer therapy. Postepy Hig Med Dosw (Online). 2017;71:997–1004.
- Anderson DB, Zanella CA, Henley JM, et al. Sumoylation: implications for neurodegenerative diseases. Adv Exp Med Biol. 2017;963:261–281.
- He X, Riceberg J, Soucy T, et al. Probing the roles of SUMOylation in cancer cell biology by using a selective SAE inhibitor. Nat Chem Biol. 2017;13(11):1164–1171.
- Matsumoto H, Saitoh H. Puromycin induces SUMO and ubiquitin redistribution upon proteasome inhibition. Biochem Biophys Res Commun. 2016;476(3):153–158.
- Liu Q, Li J, Khoury J, et al. Adenosine signaling mediates SUMO-1 modification of IkappaBalpha during hypoxia and reoxygenation. J Biol Chem. 2009;284(20):13686–13695.
- Zhao B, Villhauer EB, Bhuripanyo K, et al. SUMO-mimicking peptides inhibiting protein SUMOylation. Chembiochem. 2014;15(18):2662–2666.
- Li YJ, Du L, Wang J, et al. Allosteric inhibition of ubiquitin-like modifications by a class of inhibitor of SUMO-activating enzyme. Cell Chem Biol. 2019;26(2):278–288.
- Lv Z, Yuan L, Atkison JH, et al. Molecular mechanism of a covalent allosteric inhibitor of SUMO E1 activating enzyme. Nat Commun. 2018;9(1):5145.
- Wu J, Lei H, Zhang J, et al. Momordin Ic, a new natural SENP1 inhibitor, inhibits prostate cancer cell proliferation. Oncotarget. 2016;7(37):58995–59005.
- Ambaye N, Chen CH, Khanna S, et al. Streptonigrin inhibits SENP1 and reduces the protein level of hypoxia-inducible factor 1alpha (HIF1alpha) in cells. Biochemistry. 2018;57(11):1807–1813.
